set.seed(1)
z <- w <- rnorm(100, sd = 20)
for (t in 2:100) z[t] <- 0.8 * z[t - 1] + w[t]
Time <- 1:100
x <- 50 + 3 * Time + z
plot(x, xlab = "time", type = "l")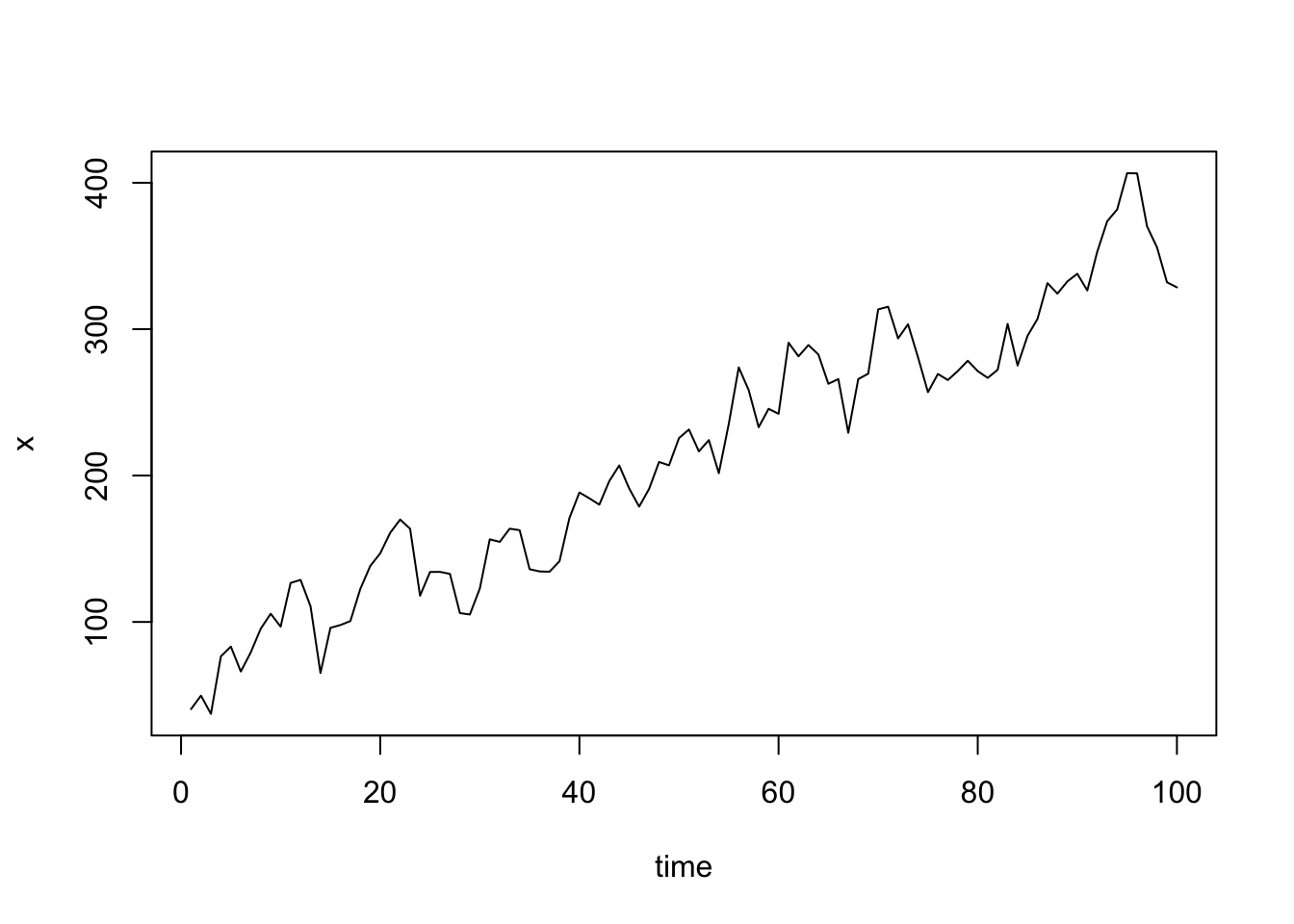
R” Modern LookR commands used in examplesIn this chapter various regression models are studied that are suitable for a time series analysis of data that contain deterministic trends and regular seasonal changes. We begin by looking at linear models for trends and then introduce regression models that account for seasonal variation using indicator and harmonic variables. Regression models can also include explanatory variables. The logarithmic transformation, which is often used to stabilise the variance, is also considered.
AIC() function and the AIC calculation in the Tidyverts output is a matter of constant terms. The general notion of reducing the value of AIC with your model still holds.R” Modern LookBook code
set.seed(1)
z <- w <- rnorm(100, sd = 20)
for (t in 2:100) z[t] <- 0.8 * z[t - 1] + w[t]
Time <- 1:100
x <- 50 + 3 * Time + z
plot(x, xlab = "time", type = "l")
Tidyverts code
pacman::p_load("tsibble", "fable", "feasts",
"tsibbledata", "fable.prophet", "tidyverse",
"patchwork")
set.seed(1)
dat <- tibble(w = rnorm(100, sd = 20)) |>
mutate(
Time = 1:n(),
z = purrr::accumulate2(
lag(w), w,
\(acc, nxt, w) 0.8 * acc + w,
.init = 0)[-1],
x = 50 + 3 * Time + z
) |>
tsibble::as_tsibble(index = Time)
autoplot(dat, .var = x)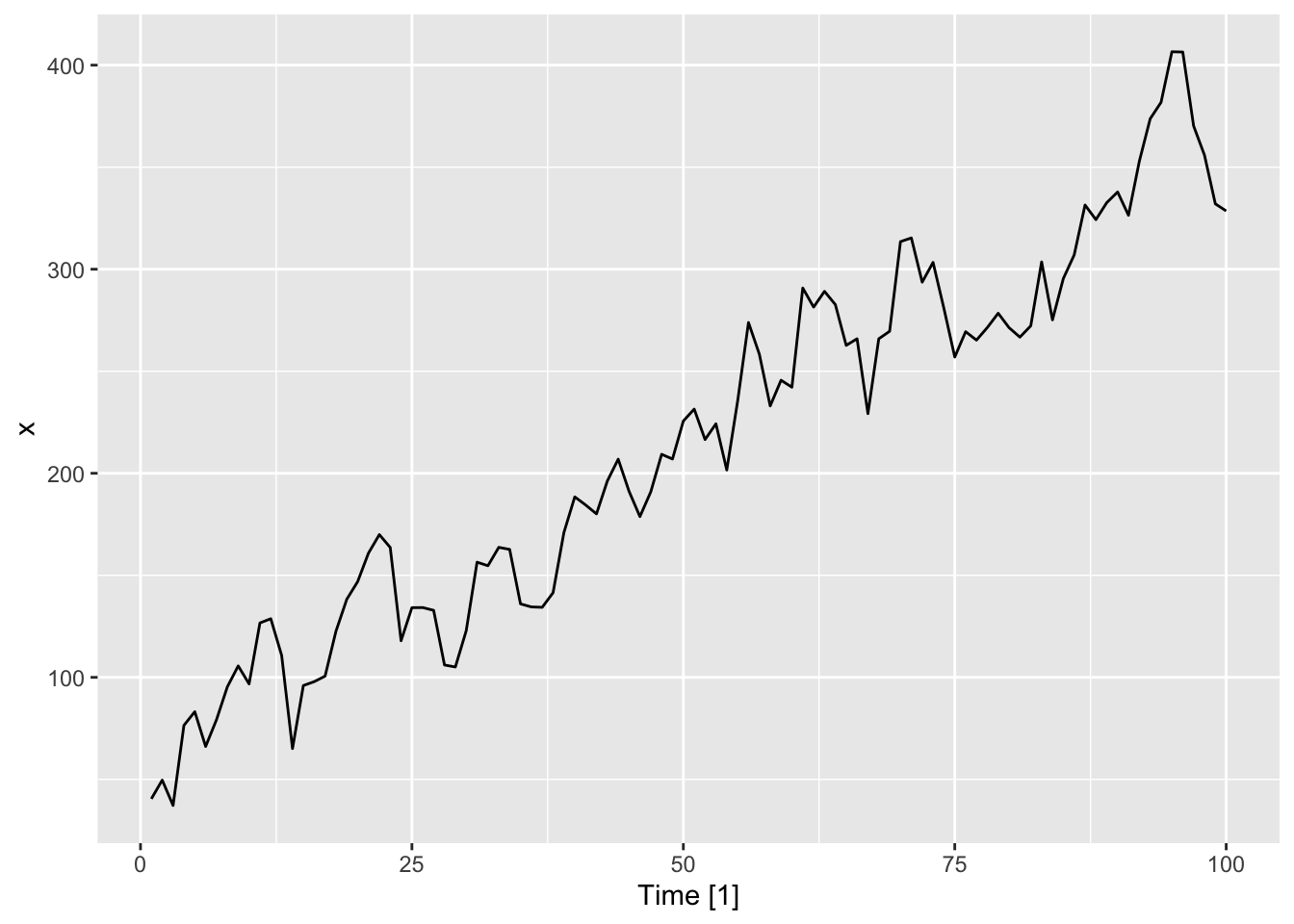
pacman::p_load("tsibble", "fable", "feasts",
"tsibbledata", "fable.prophet", "tidyverse", "patchwork")
set.seed(1)
dat <- tibble(w = rnorm(100, sd = 20)) |>
mutate(
Time = 1:n(),
z = purrr::accumulate2(
lag(w), w,
\(acc, nxt, w) 0.8 * acc + w,
.init = 0)[-1],
x = 50 + 3 * Time + z
) |>
tsibble::as_tsibble(index = Time)
autoplot(dat, .var = x)Book code
x.lm <- lm(x ~ Time)
coef(x.lm)(Intercept) Time
58.551218 3.063275 Tidyverts code
dat_lm <- dat |>
model(lm = TSLM(x ~ Time))
tidy(dat_lm) |> pull(estimate)[1] 58.551218 3.063275sqrt(diag(vcov(x.lm)))(Intercept) Time
4.88006278 0.08389621 tidy(dat_lm) |> pull(std.error)[1] 4.88006278 0.08389621acf(resid(x.lm))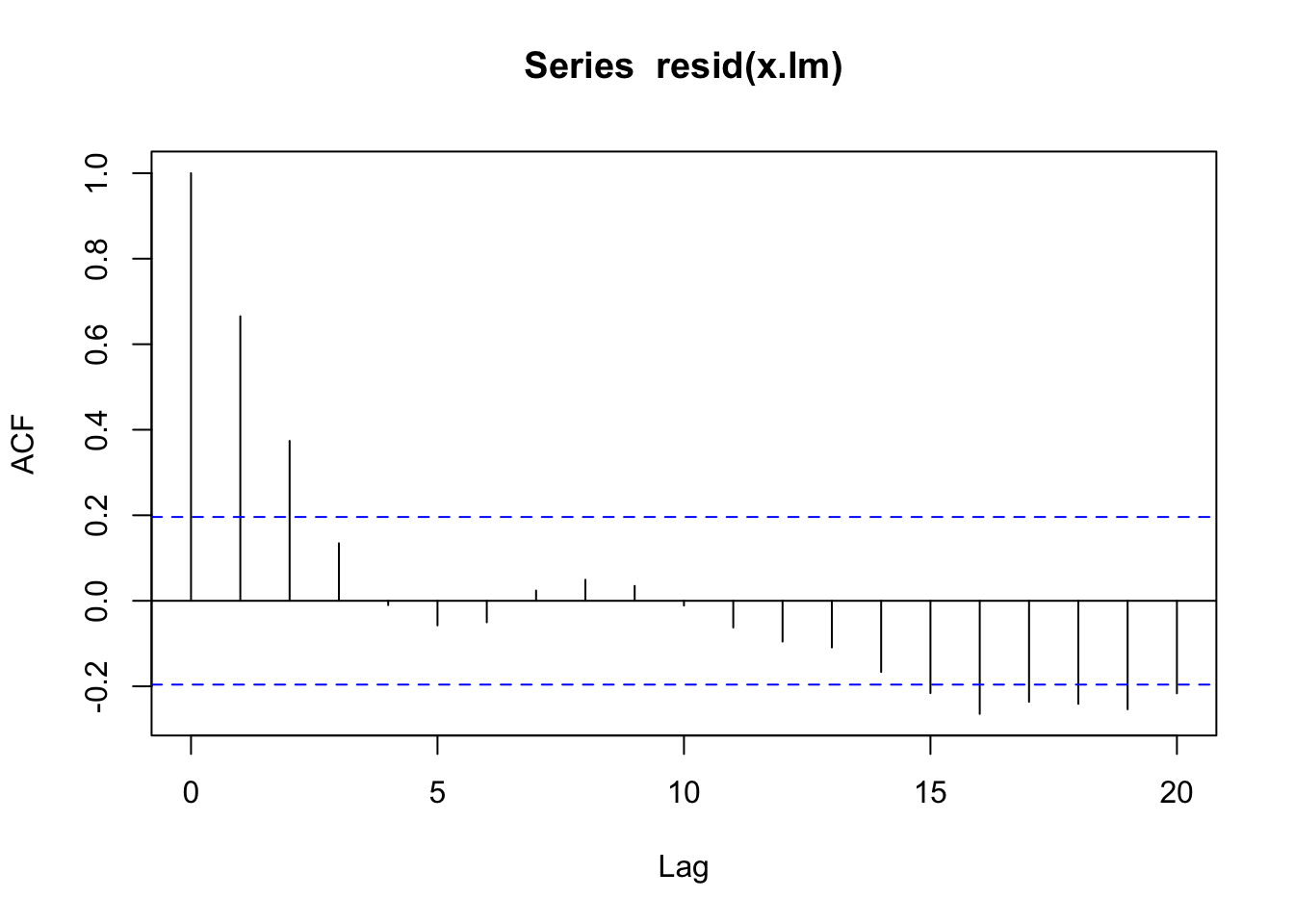
residuals(dat_lm) |> feasts::ACF(var = .resid) |> autoplot()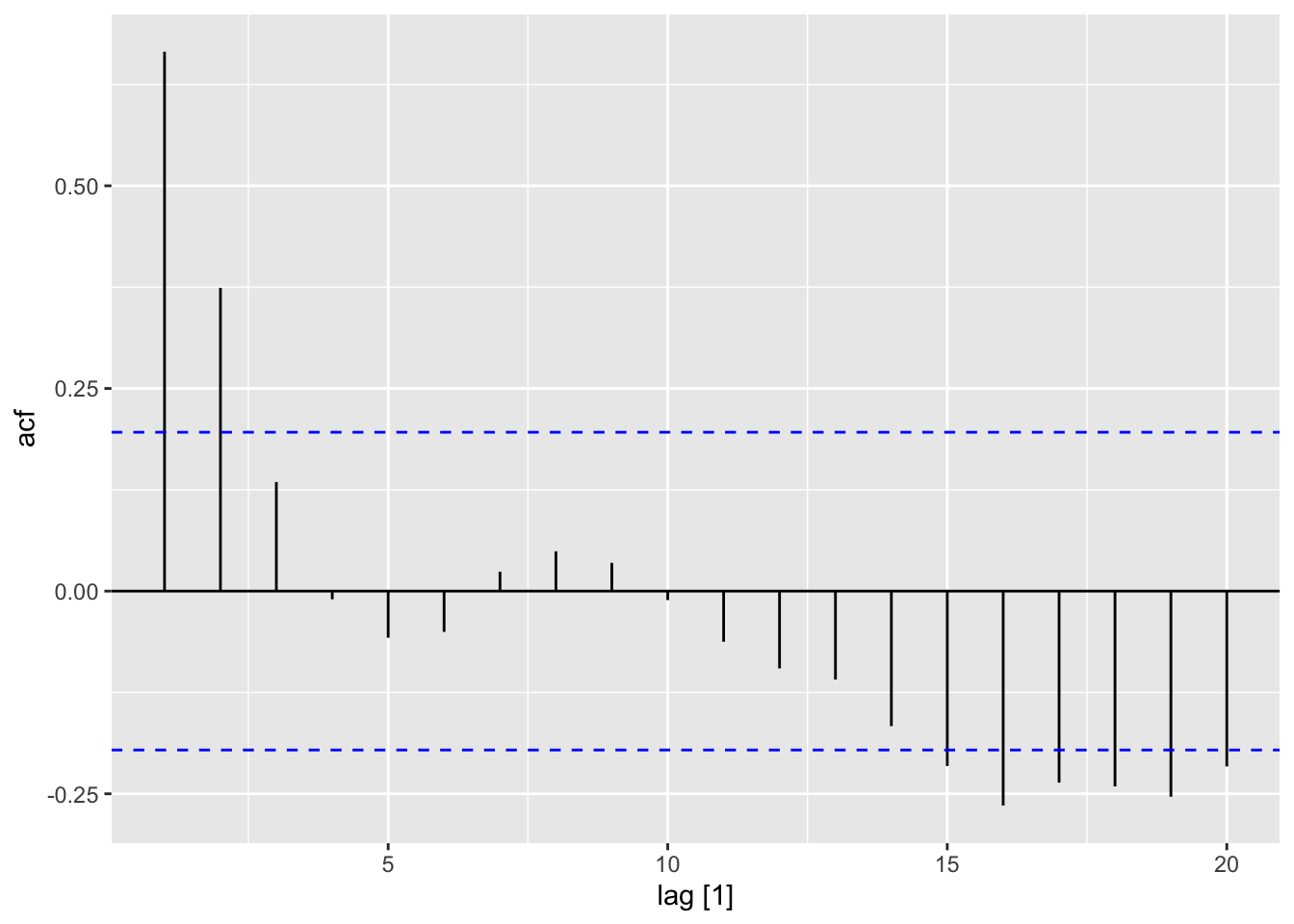
pacf(resid(x.lm))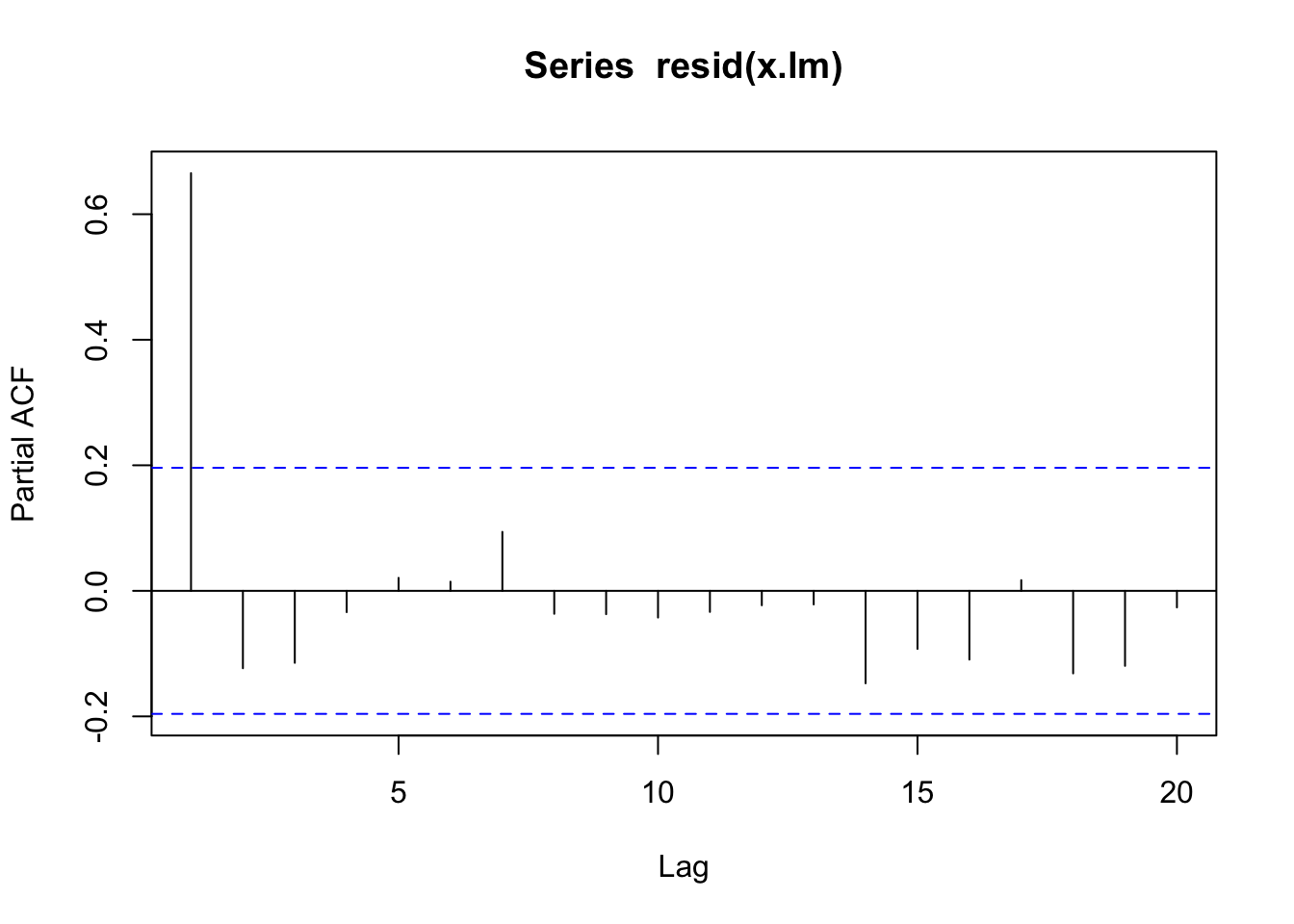
residuals(dat_lm) |> PACF(var = .resid) |> autoplot()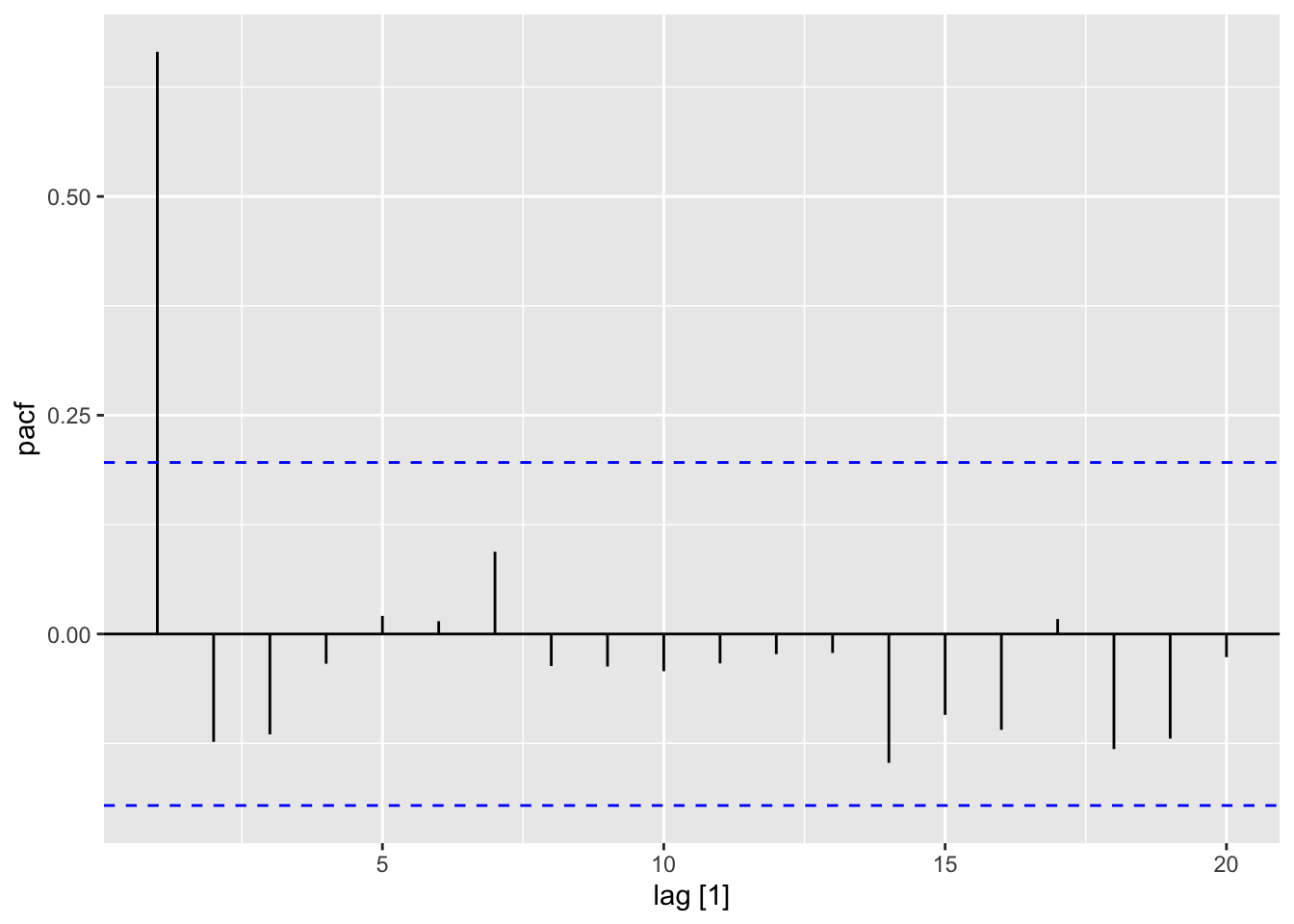
Book code
Global <- scan("data/global.dat")
Global.ts <- ts(Global, st = c(1856, 1), end = c(2005, 12), fr = 12)
temp <- window(Global.ts, start = 1970)
temp.lm <- lm(temp ~ time(temp))
coef(temp.lm)(Intercept) time(temp)
-34.920409 0.017654 Tidyverts code
global <- tibble(x = scan("data/global.dat")) |>
mutate(
date = seq(
ymd("1856-01-01"),
by = "1 months",
length.out = n()),
year = year(date),
year_month = tsibble::yearmonth(date),
stats_time = year + c(0:11)/12)
global_ts <- global |>
as_tsibble(index = year_month)
temp_tidy <- global_ts |> filter(year >= 1970)
temp_lm <- temp_tidy |>
model(lm = TSLM(x ~ stats_time ))
tidy(temp_lm) |> pull(estimate)[1] -34.920409 0.017654confint(temp.lm) 2.5 % 97.5 %
(Intercept) -37.21001248 -32.63080554
time(temp) 0.01650228 0.01880572tidy(temp_lm) |>
mutate(
`2.5%` = estimate + qnorm(.025) * std.error,
`97.5%` = estimate + qnorm(.975) * std.error
)# A tibble: 2 × 8
.model term estimate std.error statistic p.value `2.5%` `97.5%`
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 lm (Intercept) -34.9 1.16 -30.0 2.15e-107 -37.2 -32.6
2 lm stats_time 0.0177 0.000586 30.1 4.99e-108 0.0165 0.0188acf(resid(lm(temp ~ time(temp))))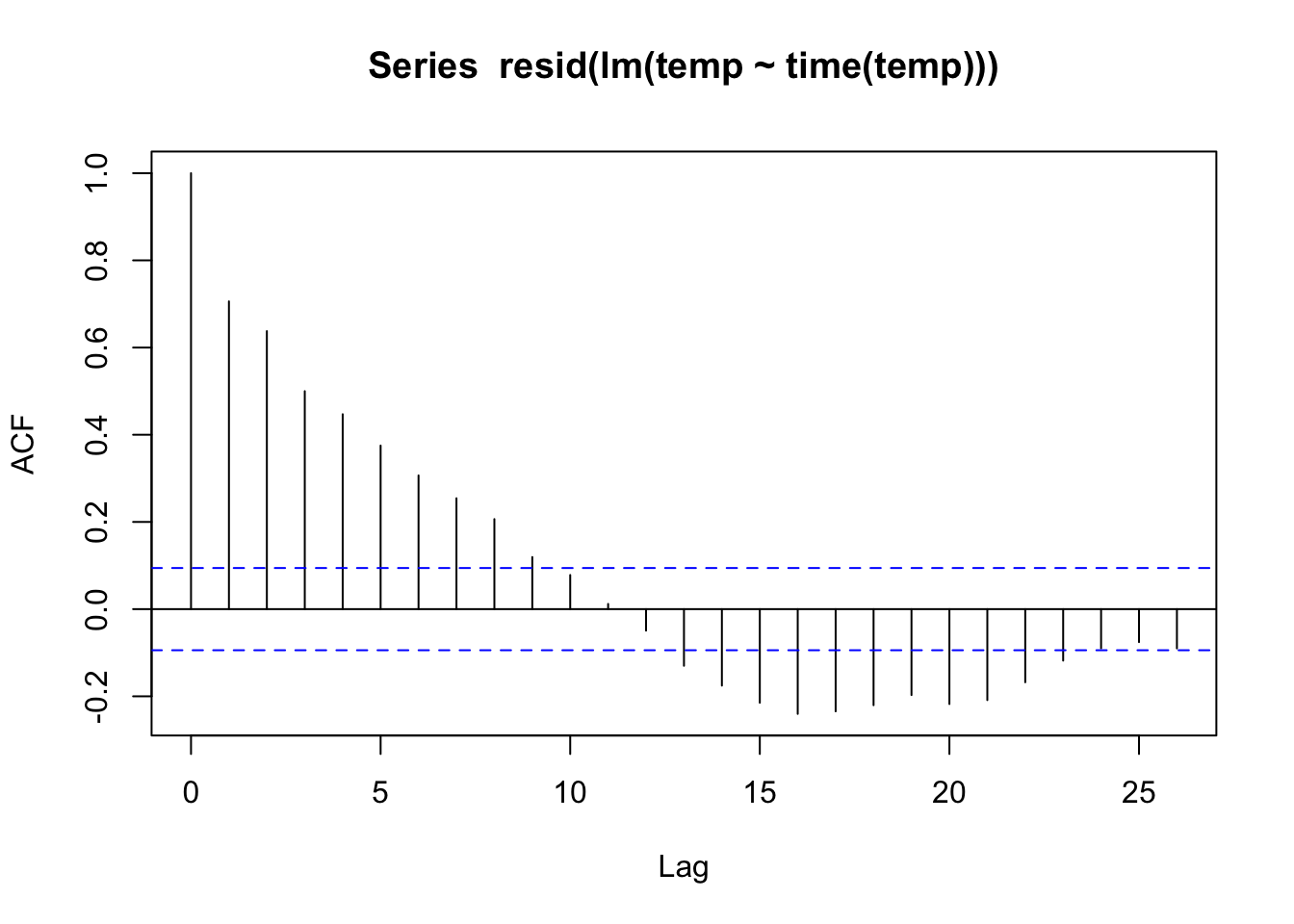
residuals(temp_lm) |>
feasts::ACF(var = .resid) |>
autoplot()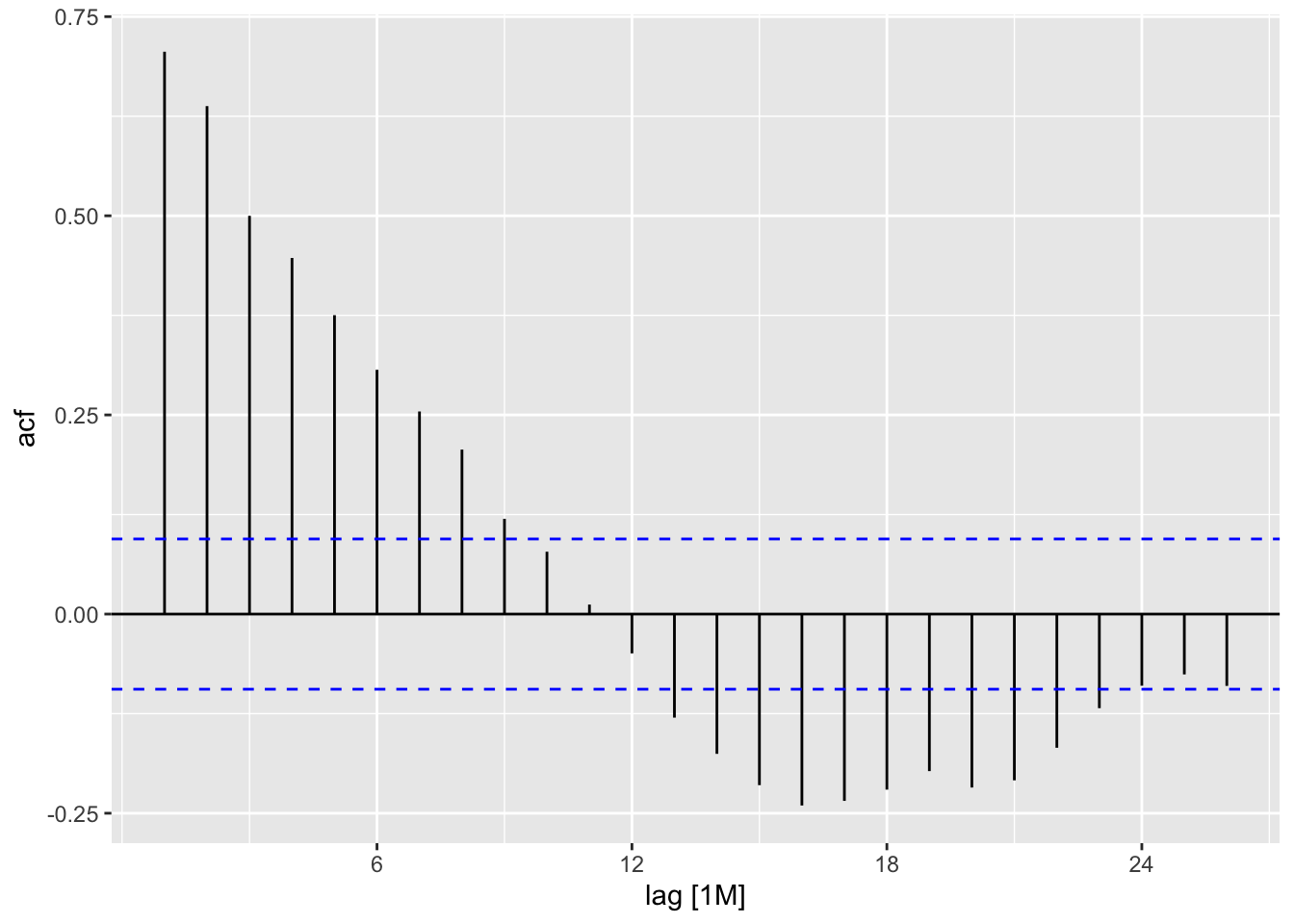
global <- tibble(x = scan("data/global.dat")) |>
mutate(
date = seq(
ymd("1856-01-01"),
by = "1 months",
length.out = n()),
year = year(date),
year_month = tsibble::yearmonth(date),
stats_time = year + c(0:11)/12)
global_ts <- global |>
as_tsibble(index = year_month)
temp_tidy <- global_ts |> filter(year >= 1970)
temp_lm <- temp_tidy |>
model(lm = TSLM(x ~ stats_time ))
tidy(temp_lm) |> pull(estimate)
tidy(temp_lm) |>
mutate(
`2.5%` = estimate + qnorm(.025) * std.error,
`97.5%` = estimate + qnorm(.975) * std.error
)
residuals(temp_lm) |>
feasts::ACF(var = .resid) |>
autoplot()Book code
library(nlme)
x.gls <- gls(x ~ Time, cor = corAR1(0.8))
coef(x.gls)(Intercept) Time
58.233018 3.042245 Tidyverts code
pacman::p_load(tidymodels, multilevelmod,
nlme, broom.mixed)
gls_spec <- linear_reg() |>
set_engine("gls", correlation = nlme::corAR1(0.8))
dat_gls <- gls_spec |>
fit(x ~ Time, data = dat)
broom.mixed::tidy(dat_gls) |>
pull(estimate)[1] 58.233018 3.042245sqrt(diag(vcov(x.gls)))(Intercept) Time
11.9245679 0.2024447 broom.mixed::tidy(dat_gls) |>
pull(std.error)[1] 11.9245679 0.2024447Book code
temp.gls <- gls(temp ~ time(temp), cor = corAR1(0.7))
confint(temp.gls) 2.5 % 97.5 %
(Intercept) -39.80571502 -28.49659113
time(temp) 0.01442275 0.02011148Tidyverts code
temp_spec <- linear_reg() |>
set_engine("gls", correlation = nlme::corAR1(0.7))
temp_gls <- temp_spec |>
fit(x ~ stats_time, data = temp_tidy)
tidy(temp_gls) |>
mutate(
`2.5%` = estimate + qnorm(.025) * std.error,
`97.5%` = estimate + qnorm(.975) * std.error
)# A tibble: 2 × 7
term estimate std.error statistic p.value `2.5%` `97.5%`
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 (Intercept) -34.2 2.89 -11.8 3.54e-28 -39.8 -28.5
2 stats_time 0.0173 0.00145 11.9 2.05e-28 0.0144 0.0201Book code
Seas <- cycle(temp)
Time <- time(temp)
temp.lm <- lm(temp ~ 0 + Time + factor(Seas))
coef(temp.lm) Time factor(Seas)1 factor(Seas)2 factor(Seas)3 factor(Seas)4
0.01770758 -34.99726483 -34.98801824 -35.01002165 -35.01227506
factor(Seas)5 factor(Seas)6 factor(Seas)7 factor(Seas)8 factor(Seas)9
-35.03369513 -35.02505965 -35.02689640 -35.02476092 -35.03831988
factor(Seas)10 factor(Seas)11 factor(Seas)12
-35.05248996 -35.06557670 -35.04871900 Tidyverts code
temp_tidy <- temp_tidy |>
mutate(month = factor(month(date)))
temp_lm <- temp_tidy |>
model(TSLM(x ~ 0 + stats_time + month))
temp_lm |>
tidy() |>
pull(estimate) [1] 0.01770758 -34.99726483 -34.98801824 -35.01002165 -35.01227506
[6] -35.03369513 -35.02505965 -35.02689640 -35.02476092 -35.03831988
[11] -35.05248996 -35.06557670 -35.04871900new.t <- seq(2006, len = 2 * 12, by = 1/12)
alpha <- coef(temp.lm)[1]
beta <- rep(coef(temp.lm)[2:13], 2)
(alpha * new.t + beta)[1:4]factor(Seas)1 factor(Seas)2 factor(Seas)3 factor(Seas)4
0.5241458 0.5348681 0.5143403 0.5135625 new_dat <- tibble(
stats_time = c(2006 + c(0:11)/12, 2007 + c(0:11)/12),
month = factor(rep(1:12, 2)),
alpha = tidy(temp_lm) |> slice(1) |> pull(estimate),
beta = rep(tidy(temp_lm) |> slice(2:13) |> pull(estimate), 2))
new_dat |>
mutate(estimate = alpha * stats_time + beta) |>
slice(1:4)# A tibble: 4 × 5
stats_time month alpha beta estimate
<dbl> <fct> <dbl> <dbl> <dbl>
1 2006 1 0.0177 -35.0 0.524
2 2006. 2 0.0177 -35.0 0.535
3 2006. 3 0.0177 -35.0 0.514
4 2006. 4 0.0177 -35.0 0.514new.dat <- data.frame(Time = new.t, Seas = rep(1:12, 2))
predict(temp.lm, new.dat)[1:24] 1 2 3 4 5 6 7 8
0.5241458 0.5348681 0.5143403 0.5135625 0.4936181 0.5037292 0.5033681 0.5069792
9 10 11 12 13 14 15 16
0.4948958 0.4822014 0.4705903 0.4889236 0.5418534 0.5525756 0.5320479 0.5312701
17 18 19 20 21 22 23 24
0.5113256 0.5214367 0.5210756 0.5246867 0.5126034 0.4999090 0.4882979 0.5066312 temp_lm |>
forecast(new_data=as_tsibble(new_dat, index = stats_time)) |>
pull(.mean) [1] 0.5241458 0.5348681 0.5143403 0.5135625 0.4936181 0.5037292 0.5033681
[8] 0.5069792 0.4948958 0.4822014 0.4705903 0.4889236 0.5418534 0.5525756
[15] 0.5320479 0.5312701 0.5113256 0.5214367 0.5210756 0.5246867 0.5126034
[22] 0.4999090 0.4882979 0.5066312Seas <- cycle(temp)
Time <- time(temp)
temp.lm <- lm(temp ~ 0 + Time + factor(Seas))
coef(temp.lm)
new.t <- seq(2006, len = 2 * 12, by = 1/12)
alpha <- coef(temp.lm)[1]
beta <- rep(coef(temp.lm)[2:13], 2)
(alpha * new.t + beta)[1:4]
new.dat <- data.frame(Time = new.t, Seas = rep(1:12, 2))
predict(temp.lm, new.dat)[1:24]new_dat <- tibble(
stats_time = c(2006 + c(0:11)/12, 2007 + c(0:11)/12),
month = factor(rep(1:12, 2)),
alpha = tidy(temp_lm) |> slice(1) |> pull(estimate),
beta = rep(tidy(temp_lm) |> slice(2:13) |> pull(estimate), 2))
new_dat |>
mutate(estimate = alpha * stats_time + beta) |>
slice(1:4)
temp_lm |>
forecast(new_data=as_tsibble(new_dat, index = stats_time)) |>
pull(.mean)Book code
TIME <- seq(1, 12, len = 1000)
plot(TIME, sin(2 * pi * TIME/12), type = "l")
Tidyverts code
dat <- tibble(
TIME = seq(1, 12, len = 1000),
y1 = sin(2 * pi * TIME/12),
y2 = sin(2 * pi * TIME/12) + 0.2 * sin(2 * pi * 2 *
TIME/12) + 0.1 * sin(2 * pi * 4 * TIME/12) + 0.1 *
cos(2 * pi * 4 * TIME/12))
ggplot(dat, aes(x = TIME, y = y1)) + geom_line()
plot(TIME, sin(2 * pi * TIME/12) + 0.2 * sin(2 * pi * 2 *
TIME/12) + 0.1 * sin(2 * pi * 4 * TIME/12) + 0.1 *
cos(2 * pi * 4 * TIME/12), type = "l")
ggplot(dat, aes(x = TIME, y = y2)) + geom_line()
dat <- tibble(
TIME = seq(1, 12, len = 1000),
y1 = sin(2 * pi * TIME/12),
y2 = sin(2 * pi * TIME/12) + 0.2 * sin(2 * pi * 2 *
TIME/12) + 0.1 * sin(2 * pi * 4 * TIME/12) + 0.1 *
cos(2 * pi * 4 * TIME/12))
ggplot(dat, aes(x = TIME, y = y1)) + geom_line()
ggplot(dat, aes(x = TIME, y = y2)) + geom_line()Book code
set.seed(1)
TIME <- 1:(10 * 12)
w <- rnorm(10 * 12, sd = 0.5)
Trend <- 0.1 + 0.005 * TIME + 0.001 * TIME^2
Seasonal <- sin(2*pi*TIME/12) + 0.2*sin(2*pi*2*TIME/12) +
0.1*sin(2*pi*4*TIME/12) + 0.1*cos(2*pi*4*TIME/12)
x <- Trend + Seasonal + w
plot(x, type = "l")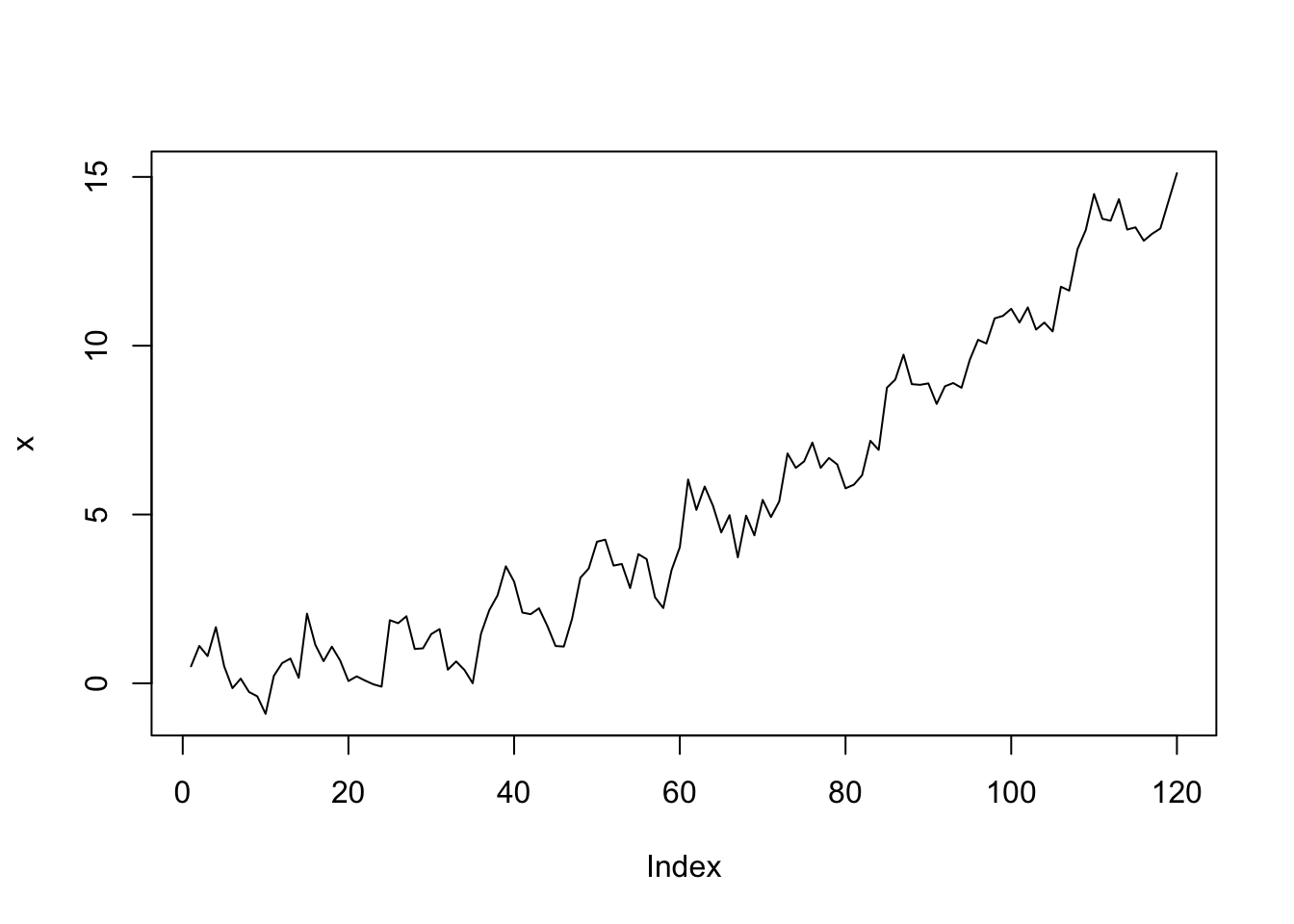
Tidyverts code
set.seed(1)
dat <- tibble(
TIME = 1:(10 * 12),
w = rnorm(10 * 12, sd = .5),
trend = 0.1 + .005 * TIME + 0.001 * TIME^2,
seasonal = sin(2 * pi * TIME / 12) +
0.2 * sin(2 * pi * 2 * TIME / 12) +
0.1 * sin(2 * pi * 4 * TIME / 12) +
0.1 * cos(2 * pi * 4 * TIME / 12),
x = trend + seasonal + w
)
ggplot(dat, aes(y = x, x = TIME)) + geom_line()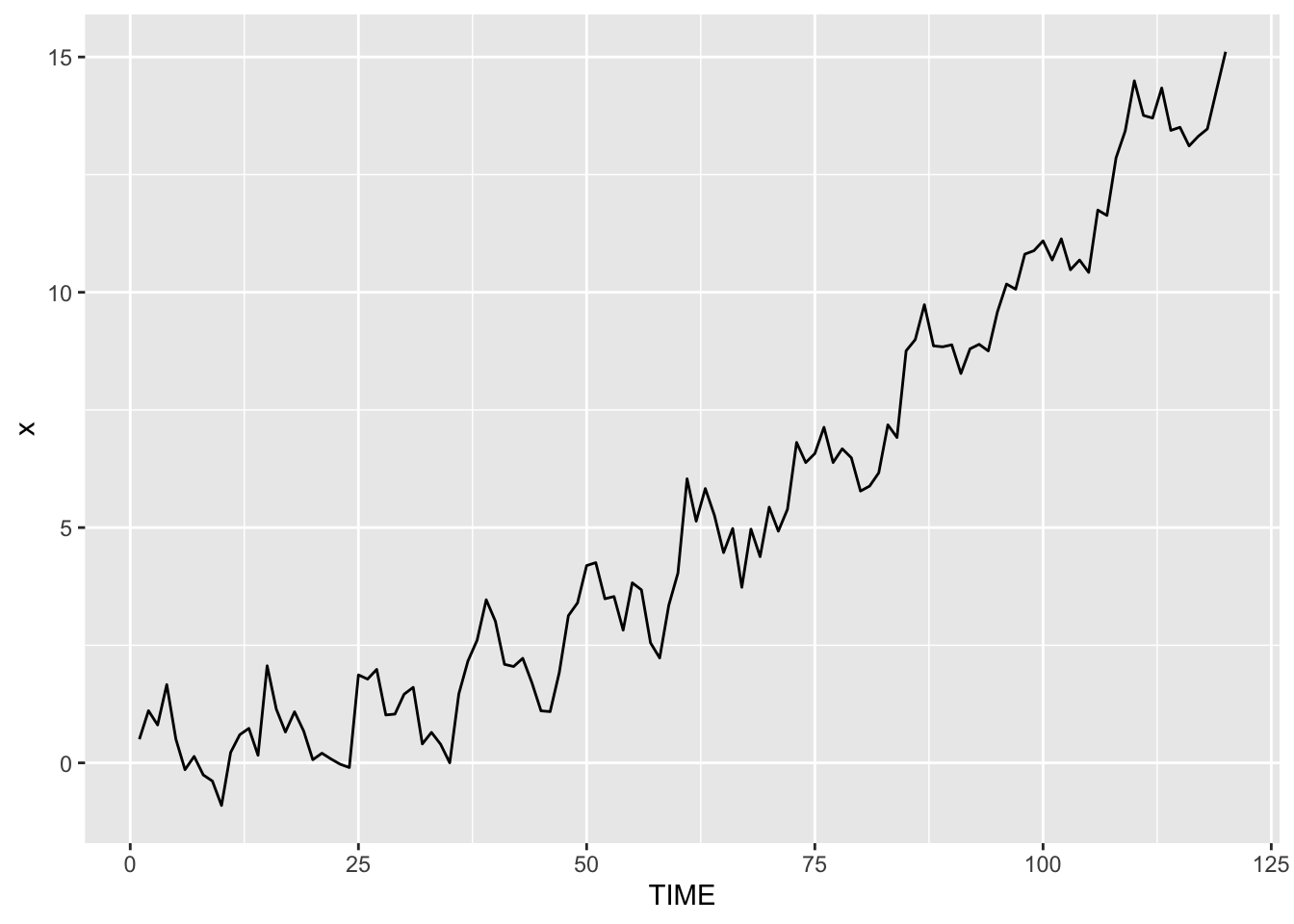
set.seed(1)
dat <- tibble(
TIME = 1:(10 * 12),
w = rnorm(10 * 12, sd = .5),
trend = 0.1 + .005 * TIME + 0.001 * TIME^2,
seasonal = sin(2 * pi * TIME / 12) +
0.2 * sin(2 * pi * 2 * TIME / 12) +
0.1 * sin(2 * pi * 4 * TIME / 12) +
0.1 * cos(2 * pi * 4 * TIME / 12),
x = trend + seasonal + w
)
ggplot(dat, aes(y = x, x = TIME)) + geom_line()Book code
SIN <- COS <- matrix(nr = length(TIME), nc = 6)
for (i in 1:6) {
COS[, i] <- cos(2 * pi * i * TIME / 12)
SIN[, i] <- sin(2 * pi * i * TIME / 12)
}
x.lm1 <- lm(x ~ TIME + I(TIME^2) + COS[, 1] + SIN[, 1] +
COS[,2] + SIN[,2] + COS[,3] + SIN[,3] + COS[,4] + SIN[, 4] +
COS[, 5] + SIN[, 5] + COS[, 6] + SIN[, 6])
coef(x.lm1) / sqrt(diag(vcov(x.lm1)))(Intercept) TIME I(TIME^2) COS[, 1] SIN[, 1] COS[, 2]
1.2390758 1.1251009 25.9327225 0.3277392 15.4421534 -0.5146340
SIN[, 2] COS[, 3] SIN[, 3] COS[, 4] SIN[, 4] COS[, 5]
3.4468638 0.2317971 -0.7027374 0.2276281 1.0525106 -1.1501059
SIN[, 5] COS[, 6] SIN[, 6]
0.8573584 -0.3100116 0.3823830 Tidyverts code
dat <- dat |>
mutate(
cos1 = cos(2 * pi * 1 * TIME/12),
cos2 = cos(2 * pi * 2 * TIME/12),
cos3 = cos(2 * pi * 3 * TIME/12),
cos4 = cos(2 * pi * 4 * TIME/12),
cos5 = cos(2 * pi * 5 * TIME/12),
cos6 = cos(2 * pi * 6 * TIME/12),
sin1 = sin(2 * pi * 1 * TIME/12),
sin2 = sin(2 * pi * 2 * TIME/12),
sin3 = sin(2 * pi * 3 * TIME/12),
sin4 = sin(2 * pi * 4 * TIME/12),
sin5 = sin(2 * pi * 5 * TIME/12),
sin6 = sin(2 * pi * 6 * TIME/12)) |>
as_tsibble(index = TIME)
dat_lm1 <- dat |>
model(full = TSLM(x ~ TIME + I(TIME^2) +
cos1 + cos2 + cos3 + cos4 + cos5 + cos6 +
sin1 + sin2 + sin3 + sin4 + sin5 + sin6))
dat_lm1 |>
tidy() |>
mutate(estimate / std.error) |>
pull(`estimate/std.error`) [1] 1.2390758 1.1251009 25.9327225 0.3277392 -0.5146340 0.2317971
[7] 0.2276281 -1.1501059 -0.3100116 15.4421534 3.4468638 -0.7027374
[13] 1.0525106 0.8573584 0.3823830x.lm2 <- lm(x ~ I(TIME^2) + SIN[, 1] + SIN[, 2])
coef(x.lm2)/sqrt(diag(vcov(x.lm2)))(Intercept) I(TIME^2) SIN[, 1] SIN[, 2]
4.627904 111.143693 15.786425 3.494708 dat_lm2 <- dat |>
model(reduced = TSLM(x ~ I(TIME^2) + sin1 + sin2))
dat_lm2 |>
tidy() |>
mutate(estimate / std.error) |>
pull(`estimate/std.error`)[1] 4.627904 111.143693 15.786425 3.494708coef(x.lm2)(Intercept) I(TIME^2) SIN[, 1] SIN[, 2]
0.280403915 0.001036409 0.900206804 0.198862866 dat_lm2 |>
tidy() |>
pull(estimate)[1] 0.280403915 0.001036409 0.900206804 0.198862866c(AIC(x.lm1), AIC(x.lm2),
AIC(lm(x ~ TIME + I(TIME^2) + SIN[,1] +
SIN[,2] +SIN[,4] +COS[,4])))[1] 165.3527 149.7256 153.0309model_three <- dat |>
model(
all = TSLM(x ~ TIME + I(TIME^2) +
cos1 + cos2 + cos3 + cos4 + cos5 + cos6 +
sin1 + sin2 + sin3 + sin4 + sin5 + sin6),
signif = TSLM(x ~ I(TIME^2) + sin1 + sin2),
actual = TSLM(x ~ TIME + I(TIME^2) + sin1 + sin2 + sin4 + cos4))
glance(model_three) |> select(.model, AIC, AICc, BIC)# A tibble: 3 × 4
.model AIC AICc BIC
<chr> <dbl> <dbl> <dbl>
1 all -175. -170. -131.
2 signif -191. -190. -177.
3 actual -188. -186. -165.SIN <- COS <- matrix(nr = length(TIME), nc = 6)
for (i in 1:6) {
COS[, i] <- cos(2 * pi * i * TIME / 12)
SIN[, i] <- sin(2 * pi * i * TIME / 12)
}
x.lm1 <- lm(x ~ TIME + I(TIME^2) + COS[, 1] + SIN[, 1] +
COS[,2] + SIN[,2] + COS[,3] + SIN[,3] + COS[,4] +
SIN[, 4] + COS[, 5] + SIN[, 5] + COS[, 6] + SIN[, 6])
coef(x.lm1) / sqrt(diag(vcov(x.lm1)))
x.lm2 <- lm(x ~ I(TIME^2) + SIN[, 1] + SIN[, 2])
coef(x.lm2)/sqrt(diag(vcov(x.lm2)))
coef(x.lm2)
AIC(x.lm1)
AIC(x.lm2)
AIC(lm(x ~ TIME + I(TIME^2) + SIN[,1] + SIN[,2] +
SIN[,4] + COS[,4]))dat <- dat |>
mutate(
cos1 = cos(2 * pi * 1 * TIME/12),
cos2 = cos(2 * pi * 2 * TIME/12),
cos3 = cos(2 * pi * 3 * TIME/12),
cos4 = cos(2 * pi * 4 * TIME/12),
cos5 = cos(2 * pi * 5 * TIME/12),
cos6 = cos(2 * pi * 6 * TIME/12),
sin1 = sin(2 * pi * 1 * TIME/12),
sin2 = sin(2 * pi * 2 * TIME/12),
sin3 = sin(2 * pi * 3 * TIME/12),
sin4 = sin(2 * pi * 4 * TIME/12),
sin5 = sin(2 * pi * 5 * TIME/12),
sin6 = sin(2 * pi * 6 * TIME/12)) |>
as_tsibble(index = TIME)
dat_lm1 <- dat |>
model(full = TSLM(x ~ TIME + I(TIME^2) +
cos1 + sin1 + cos2 + sin2 + cos3 + sin3 +
cos4 + sin4 + cos5 + cos6 + sin6))
dat_lm1 |>
tidy() |>
mutate(estimate / std.error) |>
pull(`estimate/std.error`)
dat_lm2 <- dat |>
model(reduced = TSLM(x ~ I(TIME^2) + sin1 + sin2))
dat_lm2 |>
tidy() |>
mutate(estimate / std.error) |>
pull(`estimate/std.error`)
dat_lm2 |>
tidy() |>
pull(estimate)
model_three <- dat |>
model(
full = TSLM(x ~ TIME + I(TIME^2) +
cos1 + sin1 + cos2 + sin2 + cos3 + sin3 +
cos4 + sin4 + cos5 + cos6 + sin6),
reduced = TSLM(x ~ I(TIME^2) + sin1 + sin2),
third = TSLM(x ~ TIME + I(TIME^2) + sin1 + sin2 + sin4 + cos4))
glance(model_three) |> select(.model, AIC, AICc, BIC)Note:
temp.lm1 and select(temp_lm_all, lm2)) have slightly different estimates. However, the reduced models match exactly (temp.lm2 and select(temp_lm_all, lm2)).Book code
SIN <- COS <- matrix(nr = length(temp), nc = 6)
for (i in 1:6) {
COS[, i] <- cos(2 * pi * i * time(temp))
SIN[, i] <- sin(2 * pi * i * time(temp))
}
TIME <- (time(temp) - mean(time(temp))) / sd(time(temp))
mean(time(temp))[1] 1987.958Tidyverts code
temp_tidy <- temp_tidy |>
mutate(
TIME = (stats_time - mean(stats_time)) / sd(stats_time),
cos1 = cos(2 * pi * 1 * stats_time),
cos2 = cos(2 * pi * 2 * stats_time),
cos3 = cos(2 * pi * 3 * stats_time),
cos4 = cos(2 * pi * 4 * stats_time),
cos5 = cos(2 * pi * 5 * stats_time),
cos6 = cos(2 * pi * 6 * stats_time),
sin1 = sin(2 * pi * 1 * stats_time),
sin2 = sin(2 * pi * 2 * stats_time),
sin3 = sin(2 * pi * 3 * stats_time),
sin4 = sin(2 * pi * 4 * stats_time),
sin5 = sin(2 * pi * 5 * stats_time),
sin6 = sin(2 * pi * 6 * stats_time)) |>
as_tsibble(index = stats_time)
mean(pull(temp_tidy,stats_time))[1] 1987.958sd(time(temp))[1] 10.40433sd(pull(temp_tidy, stats_time))[1] 10.40433temp.lm1 <- lm(temp ~ TIME + I(TIME^2) +
COS[,1] + SIN[,1] + COS[,2] + SIN[,2] +
COS[,3] + SIN[,3] + COS[,4] + SIN[,4] +
COS[,5] + SIN[,5] + COS[,6] + SIN[,6])
coef(temp.lm1) / sqrt(diag(vcov(temp.lm1)))(Intercept) TIME I(TIME^2) COS[, 1] SIN[, 1] COS[, 2]
18.2569117 30.2800742 1.2736899 0.7447906 2.3845512 1.2086419
SIN[, 2] COS[, 3] SIN[, 3] COS[, 4] SIN[, 4] COS[, 5]
1.9310853 0.6448116 0.3971873 0.5468025 0.1681753 0.3169782
SIN[, 5] COS[, 6] SIN[, 6]
0.3504607 -0.4498835 -0.6216650 temp_lm_all <- temp_tidy |>
model(
lm = TSLM(x ~ 0 + stats_time + month),
lm1 = TSLM(x ~ TIME + I(TIME^2) +
cos1 + sin1 + cos2 + sin2 + cos3 + sin3 +
cos4 + sin4 + cos5 + sin5 + cos6 + sin6),
lm2 = TSLM(x ~ TIME + sin1 + sin2))
temp_lm_all |> select(lm1) |> tidy() |>
mutate(calc = estimate / std.error) |>
select(term, calc)# A tibble: 15 × 2
term calc
<chr> <dbl>
1 (Intercept) 18.2
2 TIME 30.2
3 I(TIME^2) 1.28
4 cos1 0.760
5 sin1 2.37
6 cos2 1.38
7 sin2 1.62
8 cos3 0.644
9 sin3 0.391
10 cos4 0.545
11 sin4 0.170
12 cos5 0.320
13 sin5 0.355
14 cos6 -0.193
15 sin6 0.0608temp.lm2 <- lm(temp ~ TIME + SIN[, 1] + SIN[, 2])
coef(temp.lm2)(Intercept) TIME SIN[, 1] SIN[, 2]
0.17501157 0.18409618 0.02041803 0.01615374 temp_lm_all |> select(lm2) |> tidy() |>
select(term, estimate)# A tibble: 4 × 2
term estimate
<chr> <dbl>
1 (Intercept) 0.175
2 TIME 0.184
3 sin1 0.0204
4 sin2 0.0162c(AIC(temp.lm), AIC(temp.lm1), AIC(temp.lm2))[1] -546.9577 -545.0491 -561.1779glance(temp_lm_all) |> select(.model, AIC, AICc, BIC)# A tibble: 3 × 4
.model AIC AICc BIC
<chr> <dbl> <dbl> <dbl>
1 lm -1773. -1772. -1716.
2 lm1 -1771. -1769. -1706.
3 lm2 -1787. -1787. -1767.plot(time(temp), resid(temp.lm2), type = "l")
abline(0, 0, col = "red")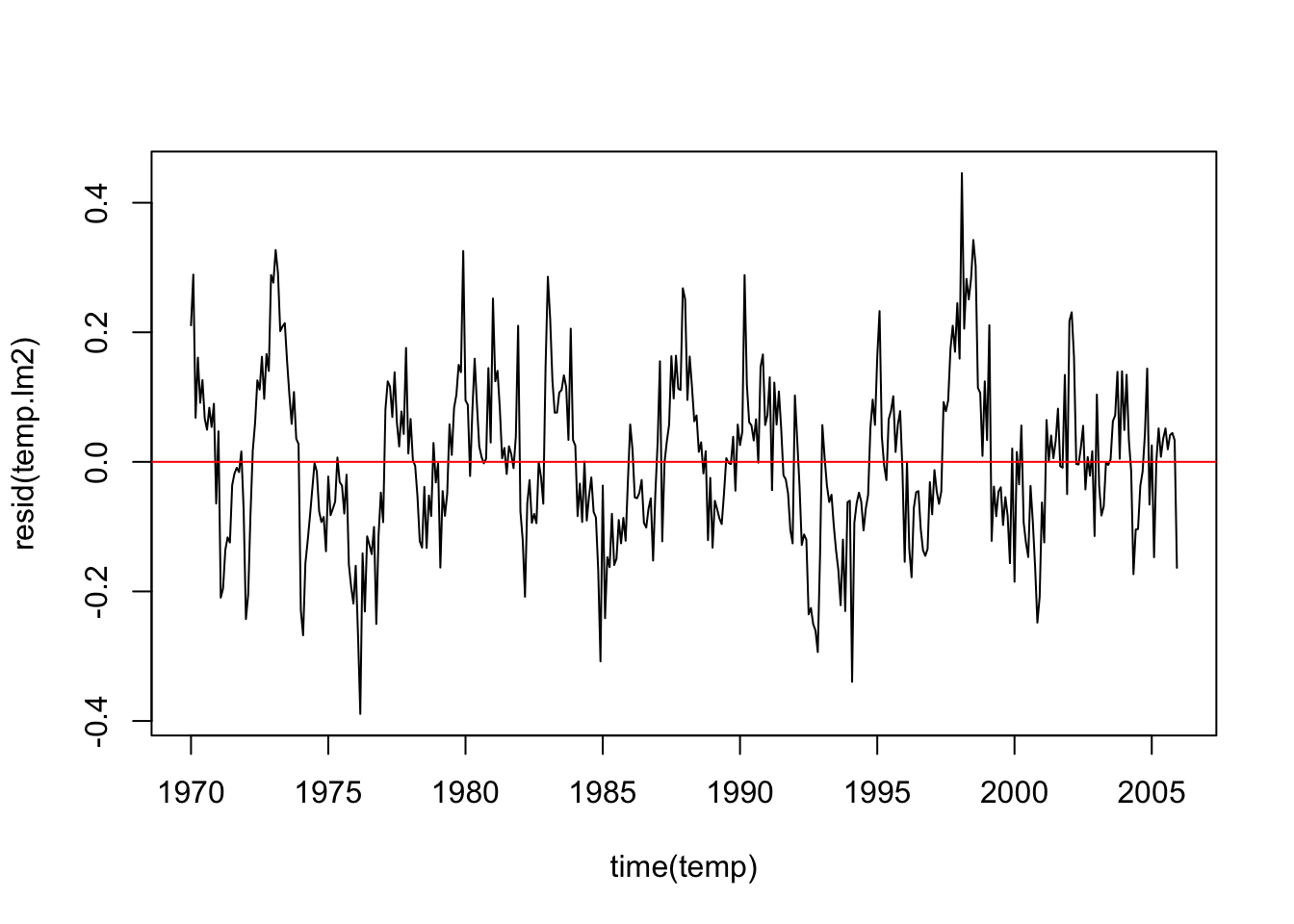
lm2_res <- temp_lm_all |>
select(lm2) |>
residuals()
lm2_res |>
autoplot(.vars = .resid) +
geom_hline(yintercept = 0, color = "red")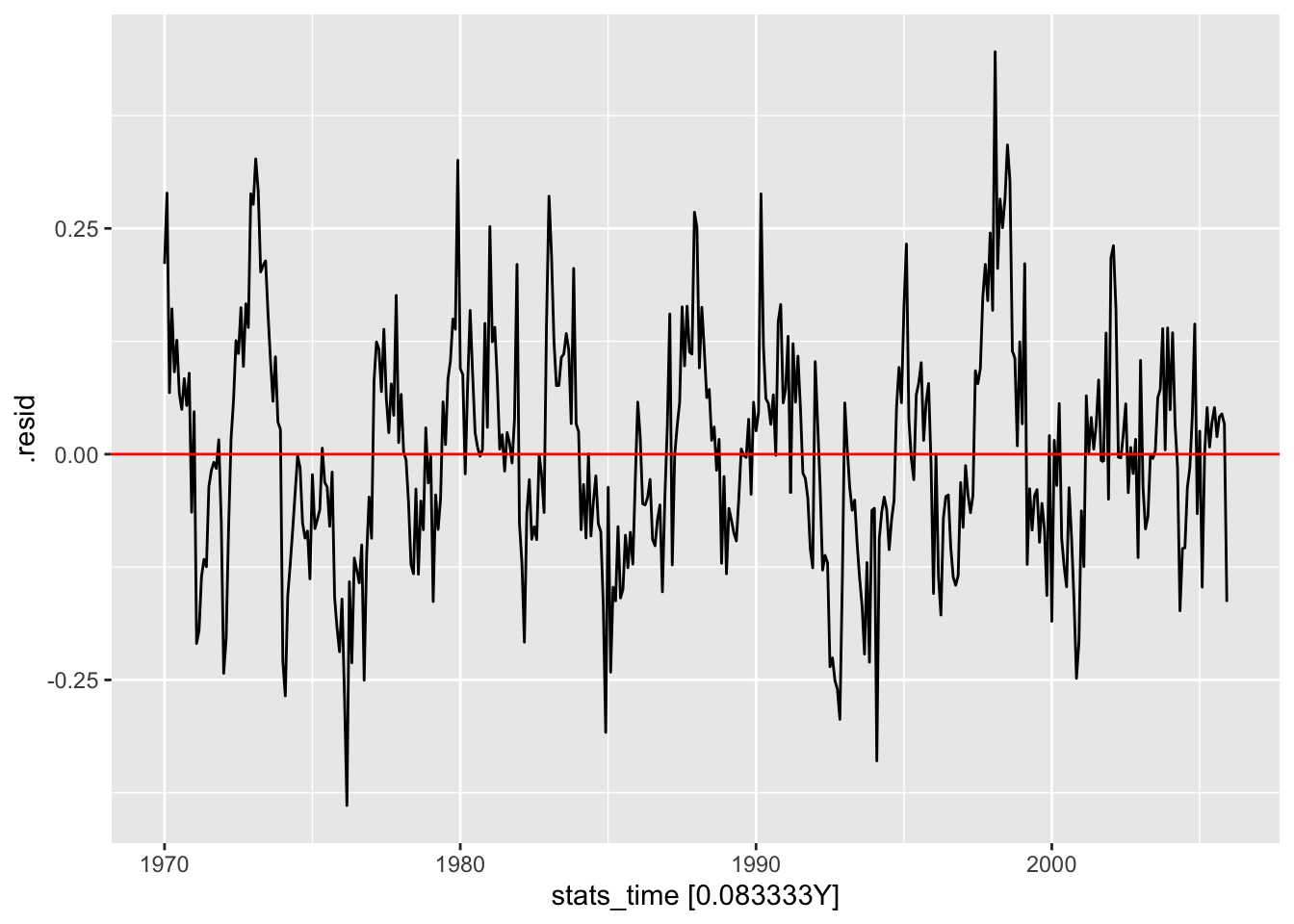
acf(resid(temp.lm2))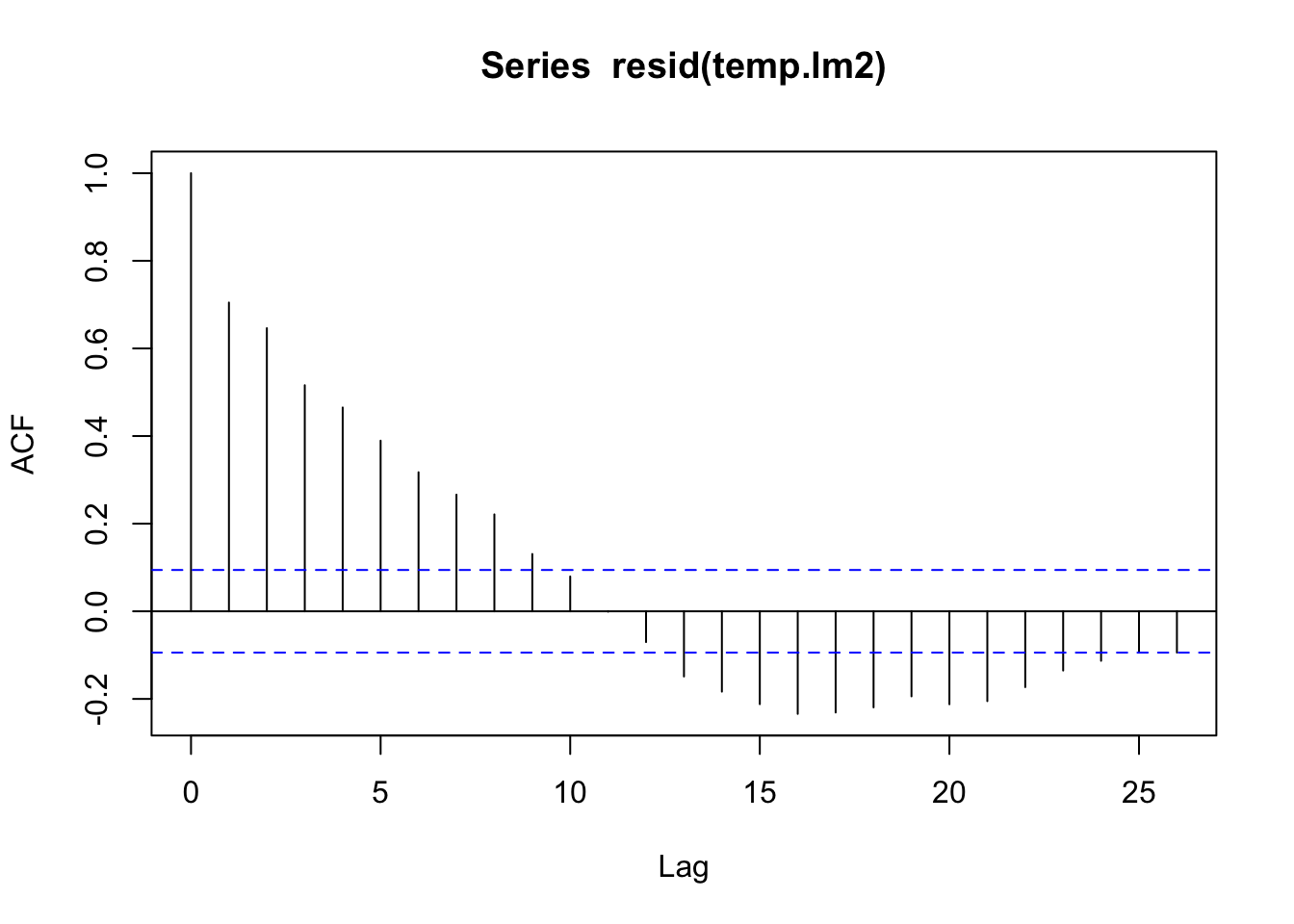
lm2_res |>
feasts::ACF(var = .resid) |>
autoplot()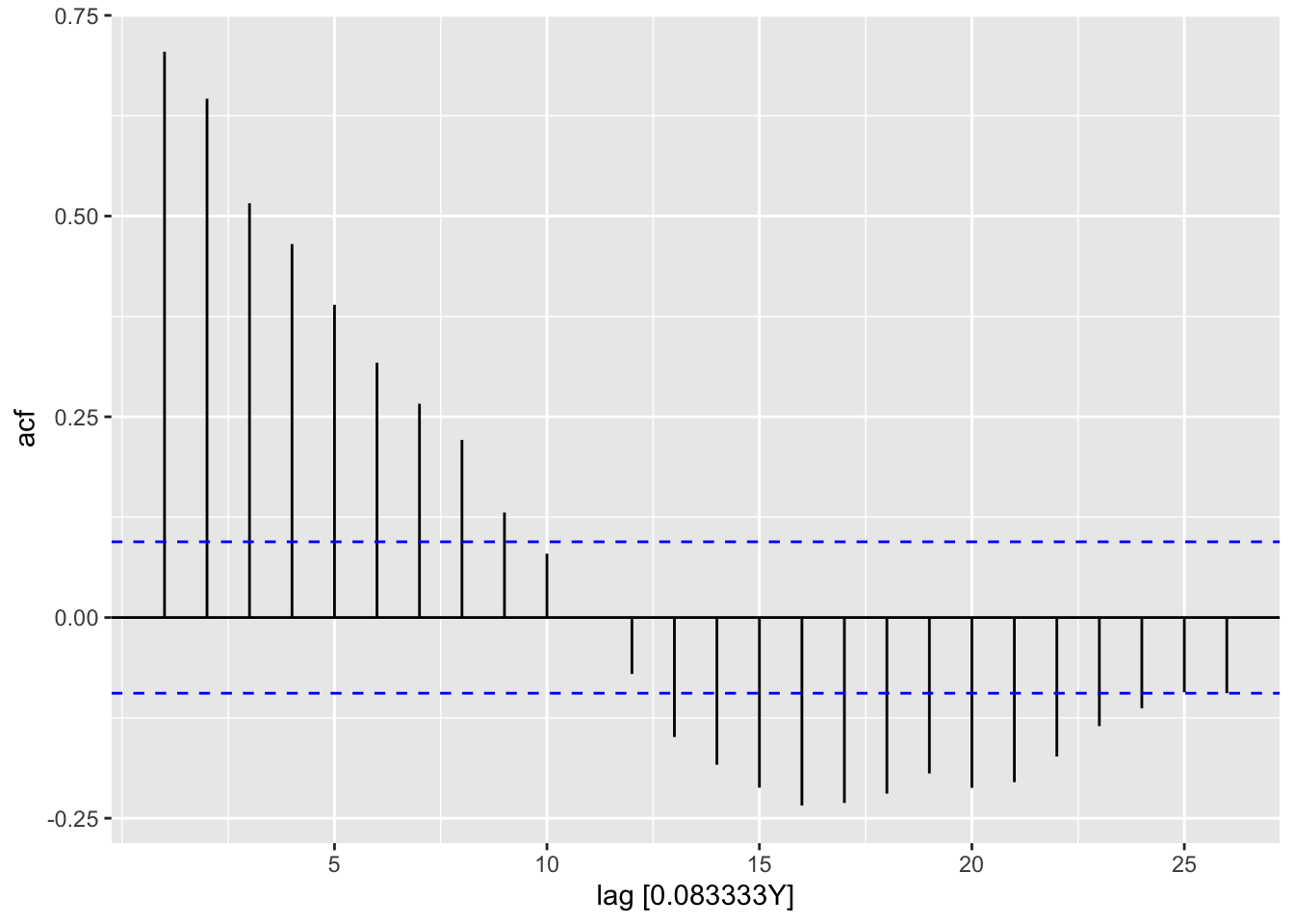
pacf(resid(temp.lm2))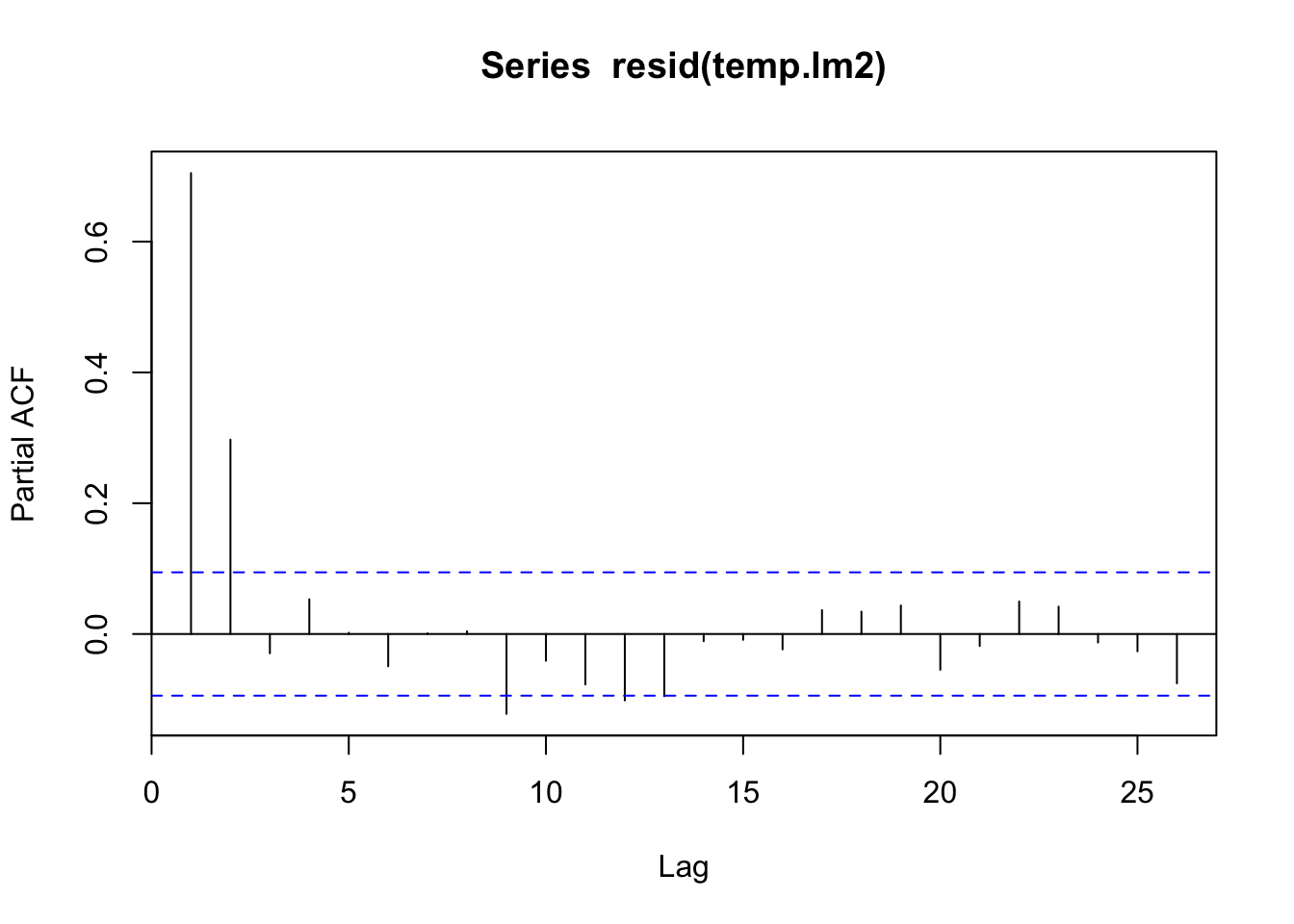
lm2_res |>
feasts::PACF(var = .resid) |>
autoplot()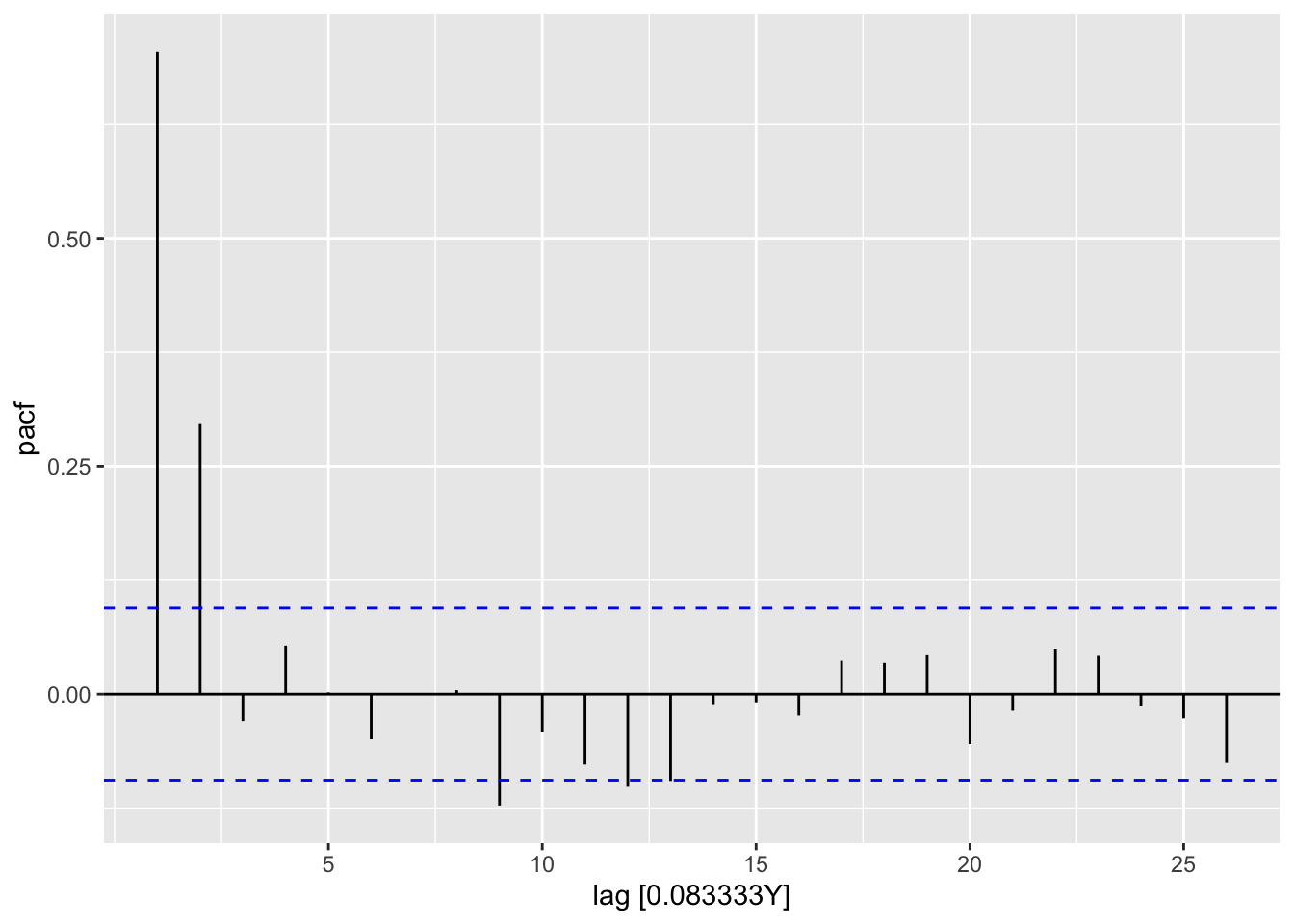
res.ar <- ar(resid(temp.lm2), method = "mle")
res.ar$ar[1] 0.4938189 0.3071598res_ar <- lm2_res |>
select(-.model) |>
model(resid = AR(.resid ~ order(1:2)))
tidy(res_ar)# A tibble: 2 × 6
.model term estimate std.error statistic p.value
<chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 resid ar1 0.486 0.0460 10.6 2.64e-23
2 resid ar2 0.305 0.0459 6.64 9.30e-11sd(res.ar$res[-(1:2)])[1] 0.08373324sd(pull(residuals(res_ar), .resid), na.rm = TRUE)[1] 0.08372416acf(res.ar$res[-(1:2)])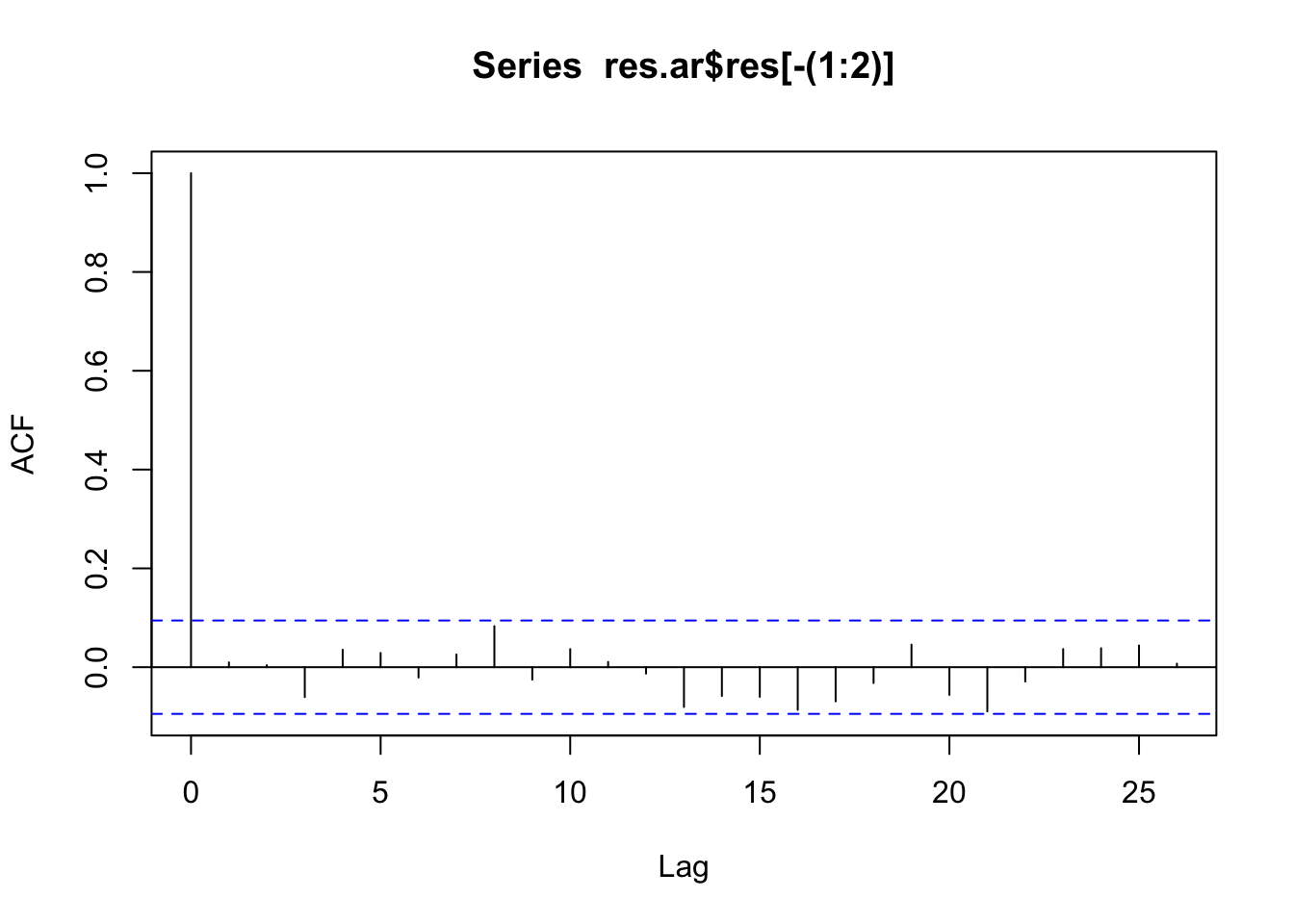
residuals(res_ar) |>
feasts::ACF(var = .resid) |>
autoplot()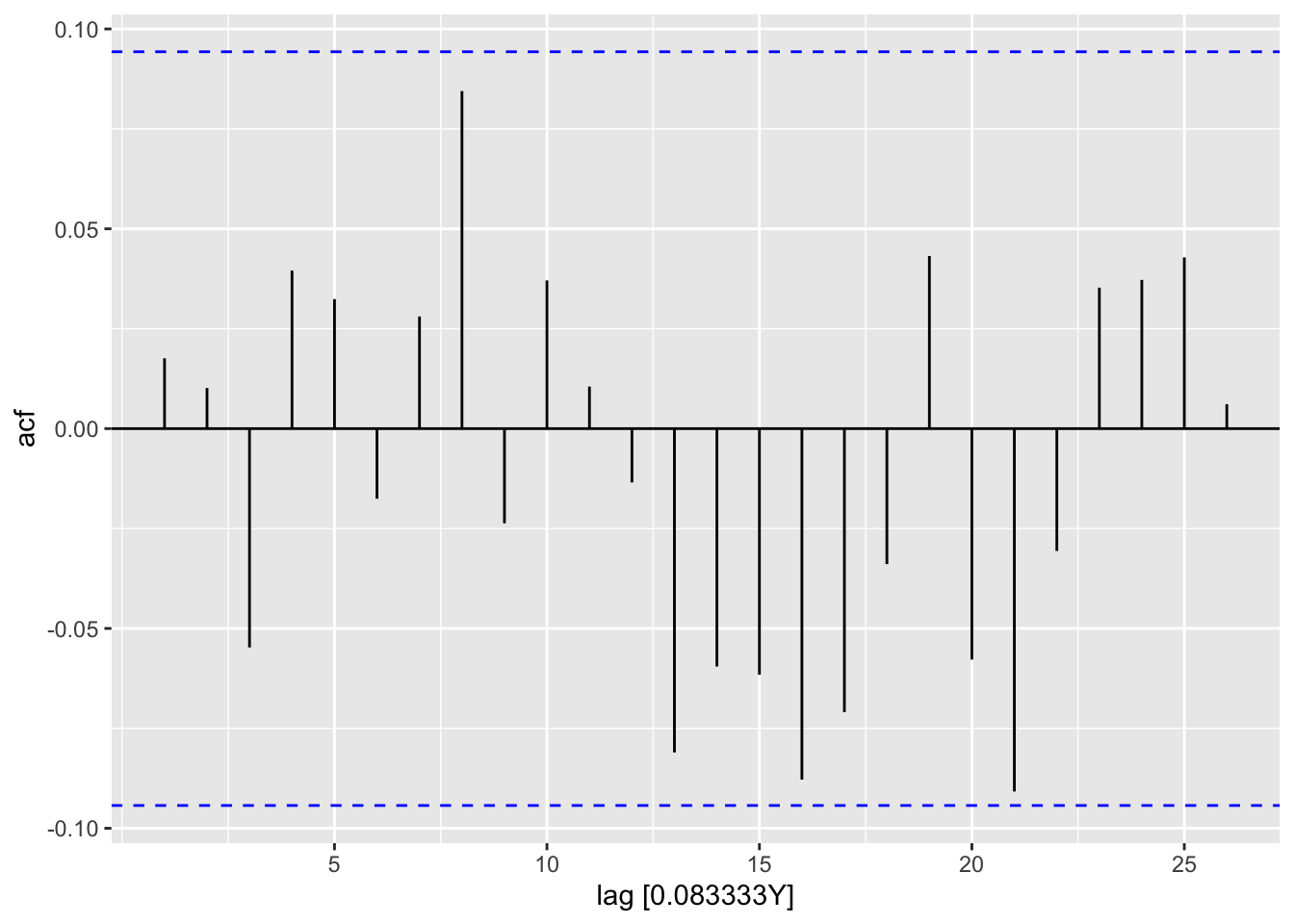
SIN <- COS <- matrix(nr = length(temp), nc = 6)
for (i in 1:6) {
COS[, i] <- cos(2 * pi * i * time(temp))
SIN[, i] <- sin(2 * pi * i * time(temp))
}
TIME <- (time(temp) - mean(time(temp))) / sd(time(temp))
mean(time(temp))
sd(time(temp))
temp.lm1 <- lm(temp ~ TIME + I(TIME^2) +
COS[,1] + SIN[,1] + COS[,2] + SIN[,2] +
COS[,3] + SIN[,3] + COS[,4] + SIN[,4] +
COS[,5] + SIN[,5] + COS[,6] + SIN[,6])
temp.lm2 <- lm(temp ~ TIME + SIN[, 1] + SIN[, 2])
coef(temp.lm1)/sqrt(diag(vcov(temp.lm1)))
coef(temp.lm2)
AIC(temp.lm)
AIC(temp.lm1)
AIC(temp.lm2)
plot(time(temp), resid(temp.lm2), type = "l")
abline(0, 0, col = "red")
acf(resid(temp.lm2))
pacf(resid(temp.lm2))
res.ar <- ar(resid(temp.lm2), method = "mle")
res.ar$ar
sd(res.ar$res[-(1:2)])
acf(res.ar$res[-(1:2)])temp_tidy <- temp_tidy |>
mutate(
TIME = (stats_time - mean(stats_time)) / sd(stats_time),
cos1 = cos(2 * pi * 1 * stats_time),
cos2 = cos(2 * pi * 2 * stats_time),
cos3 = cos(2 * pi * 3 * stats_time),
cos4 = cos(2 * pi * 4 * stats_time),
cos5 = cos(2 * pi * 5 * stats_time),
cos6 = cos(2 * pi * 6 * stats_time),
sin1 = sin(2 * pi * 1 * stats_time),
sin2 = sin(2 * pi * 2 * stats_time),
sin3 = sin(2 * pi * 3 * stats_time),
sin4 = sin(2 * pi * 4 * stats_time),
sin5 = sin(2 * pi * 5 * stats_time),
sin6 = sin(2 * pi * 6 * stats_time)) |>
as_tsibble(index = stats_time)
mean(pull(temp_tidy,stats_time))
sd(pull(temp_tidy, stats_time))
temp_lm_all <- temp_tidy |>
model(
lm = TSLM(x ~ 0 + stats_time + month),
lm1 = TSLM(x ~ TIME + I(TIME^2) +
cos1 + cos2 + cos3 + cos4 + cos5 + cos6 +
sin1 + sin2 + sin3 + sin4 + sin5 + sin6),
lm2 = TSLM(x ~ TIME + sin1 + sin2))
temp_lm_all |> select(lm1) |> tidy() |>
mutate(calc = estimate / std.error) |>
select(term, calc)
glance(temp_lm_all) |> select(.model, AIC, AICc, BIC)
lm2_res <- temp_lm_all |>
select(lm2) |>
residuals()
lm2_res |>
autoplot(.vars = .resid) +
geom_hline(yintercept = 0, color = "red")
lm2_res |>
feasts::ACF(var = .resid) |>
autoplot()
lm2_res |>
feasts::PACF(var = .resid) |>
autoplot()
res_ar <- lm2_res |>
select(-.model) |>
model(resid = AR(.resid ~ order(1:2)))
tidy(res_ar)
sd(pull(residuals(res_ar), .resid), na.rm = TRUE)
residuals(res_ar) |>
feasts::ACF(var = .resid) |>
autoplot()NOTE:
AP.lm1 and select(ap_lm, lm1)) have slightly different estimates. However, the reduced models match exactly (AP.lm2 and select(ap_lm, lm2)).gls models is different. One fits with a mean, and one fits without a mean. Additionally, one is using OLS, and the other is using MLE.Book code
data(AirPassengers)
AP <- AirPassengers
SIN <- COS <- matrix(nr = length(AP), nc = 6)
for (i in 1:6) {
SIN[, i] <- sin(2 * pi * i * time(AP))
COS[, i] <- cos(2 * pi * i * time(AP))
}
TIME <- (time(AP) - mean(time(AP)))/sd(time(AP))
plot(AP)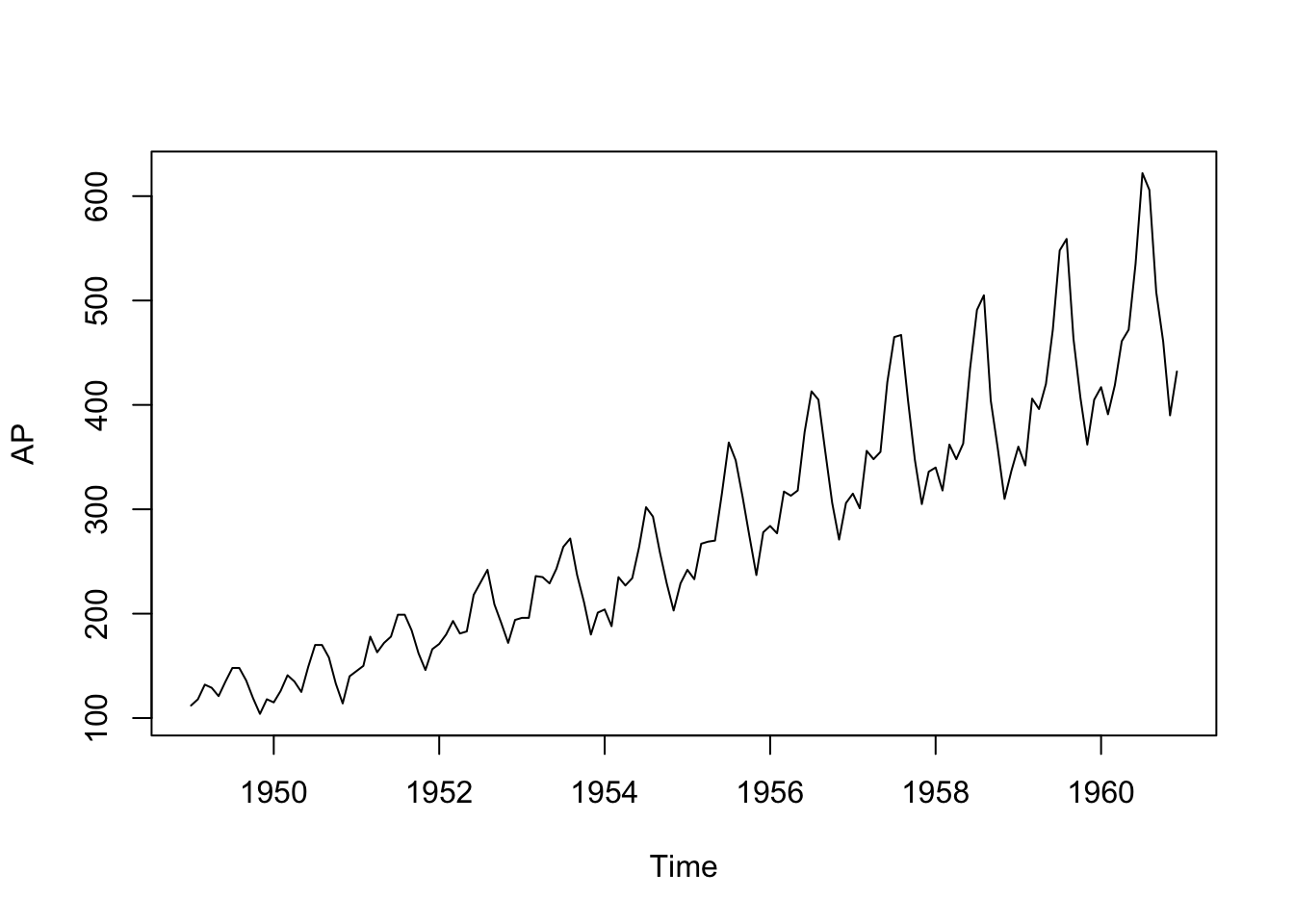
Tidyverts code
data(AirPassengers)
ap <- as_tsibble(AirPassengers) |>
mutate(
Year = year(index),
Month = month(index),
stats_time = Year + (Month - 1)/12,
TIME = (stats_time - mean(stats_time)) / sd(stats_time),
cos1 = cos(2 * pi * 1 * stats_time),
cos2 = cos(2 * pi * 2 * stats_time),
cos3 = cos(2 * pi * 3 * stats_time),
cos4 = cos(2 * pi * 4 * stats_time),
cos5 = cos(2 * pi * 5 * stats_time),
cos6 = cos(2 * pi * 6 * stats_time),
sin1 = sin(2 * pi * 1 * stats_time),
sin2 = sin(2 * pi * 2 * stats_time),
sin3 = sin(2 * pi * 3 * stats_time),
sin4 = sin(2 * pi * 4 * stats_time),
sin5 = sin(2 * pi * 5 * stats_time),
sin6 = sin(2 * pi * 6 * stats_time)) |>
as_tsibble(index = stats_time)
autoplot(ap)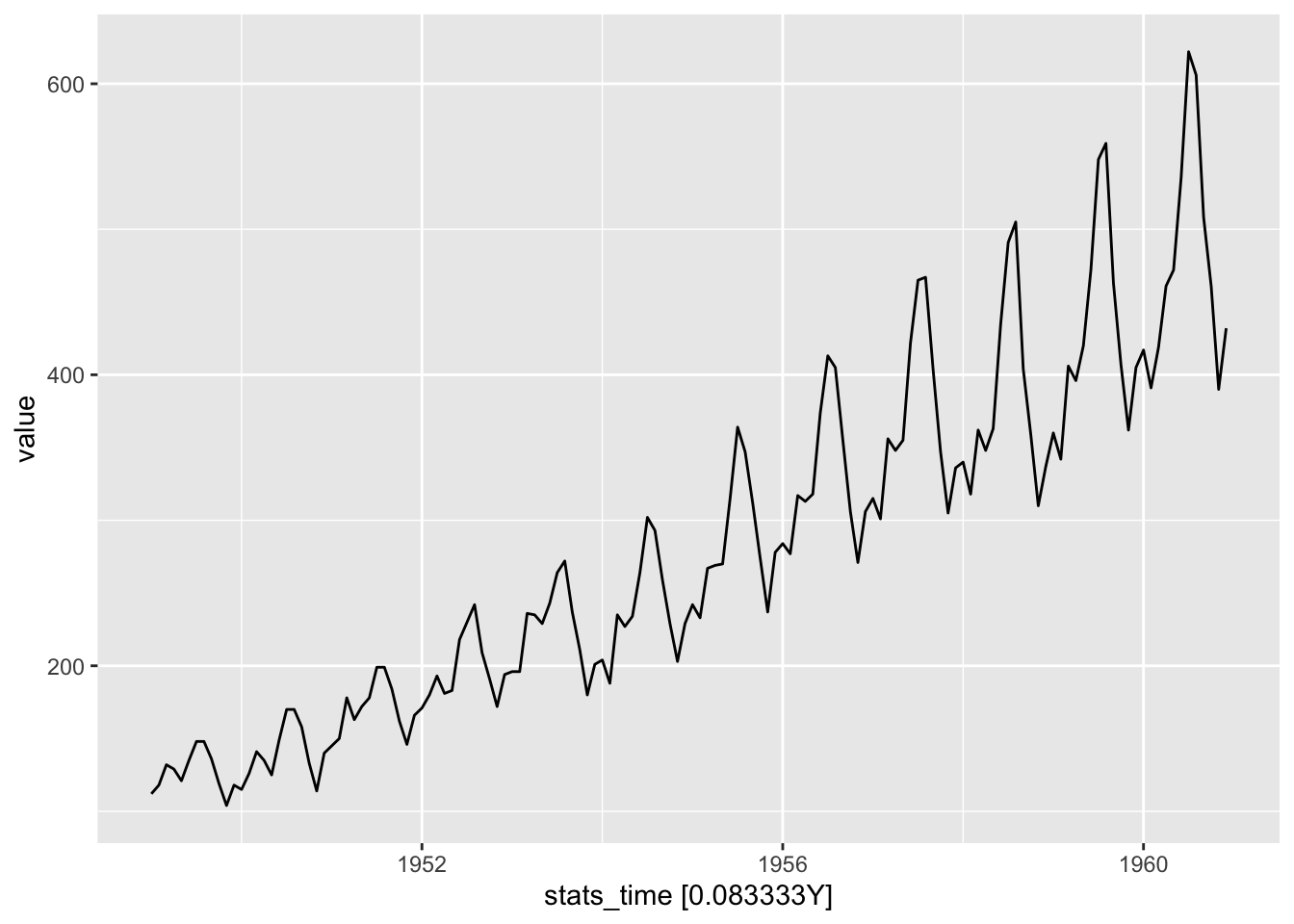
plot(log(AP))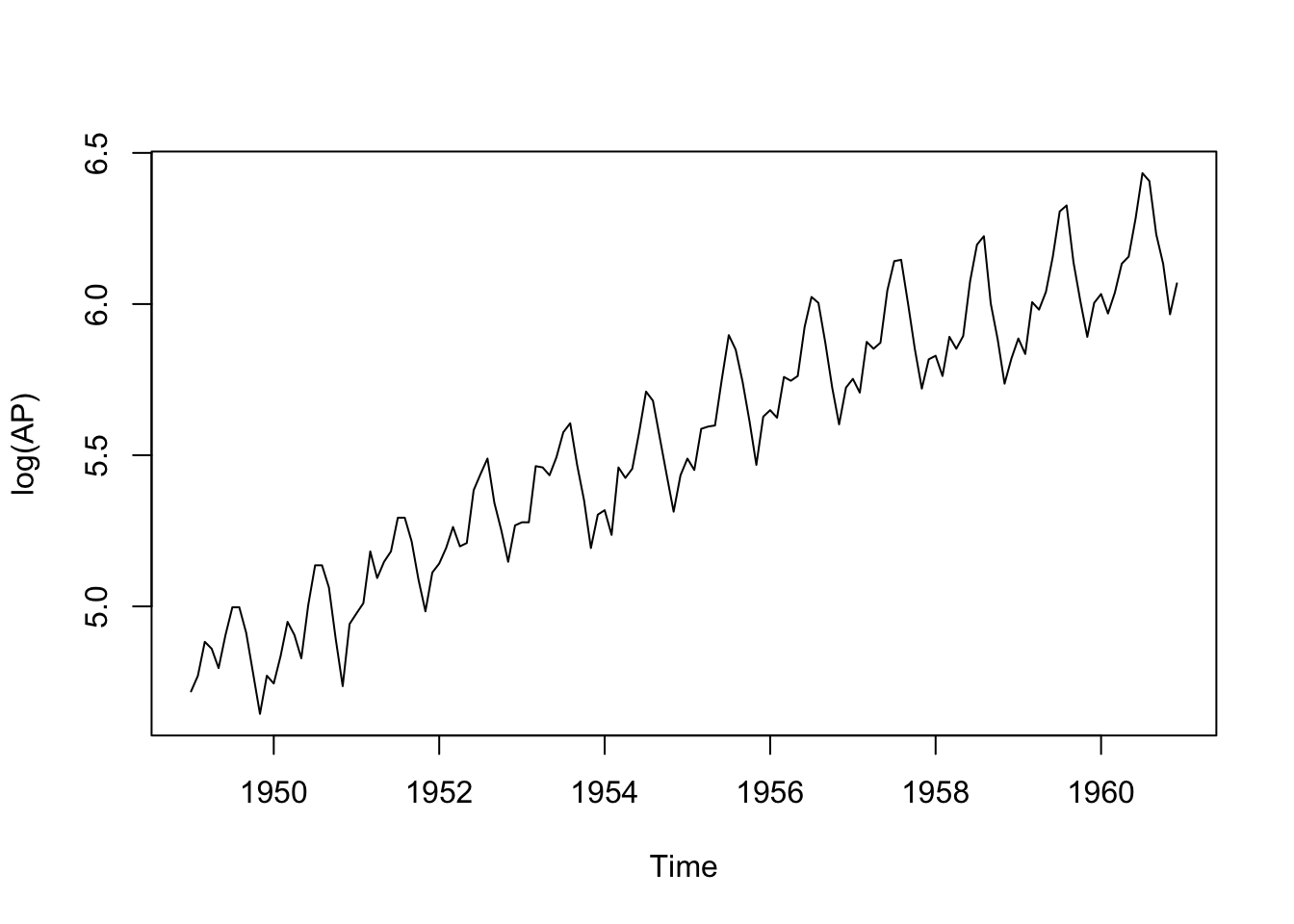
autoplot(ap, .vars = log(value))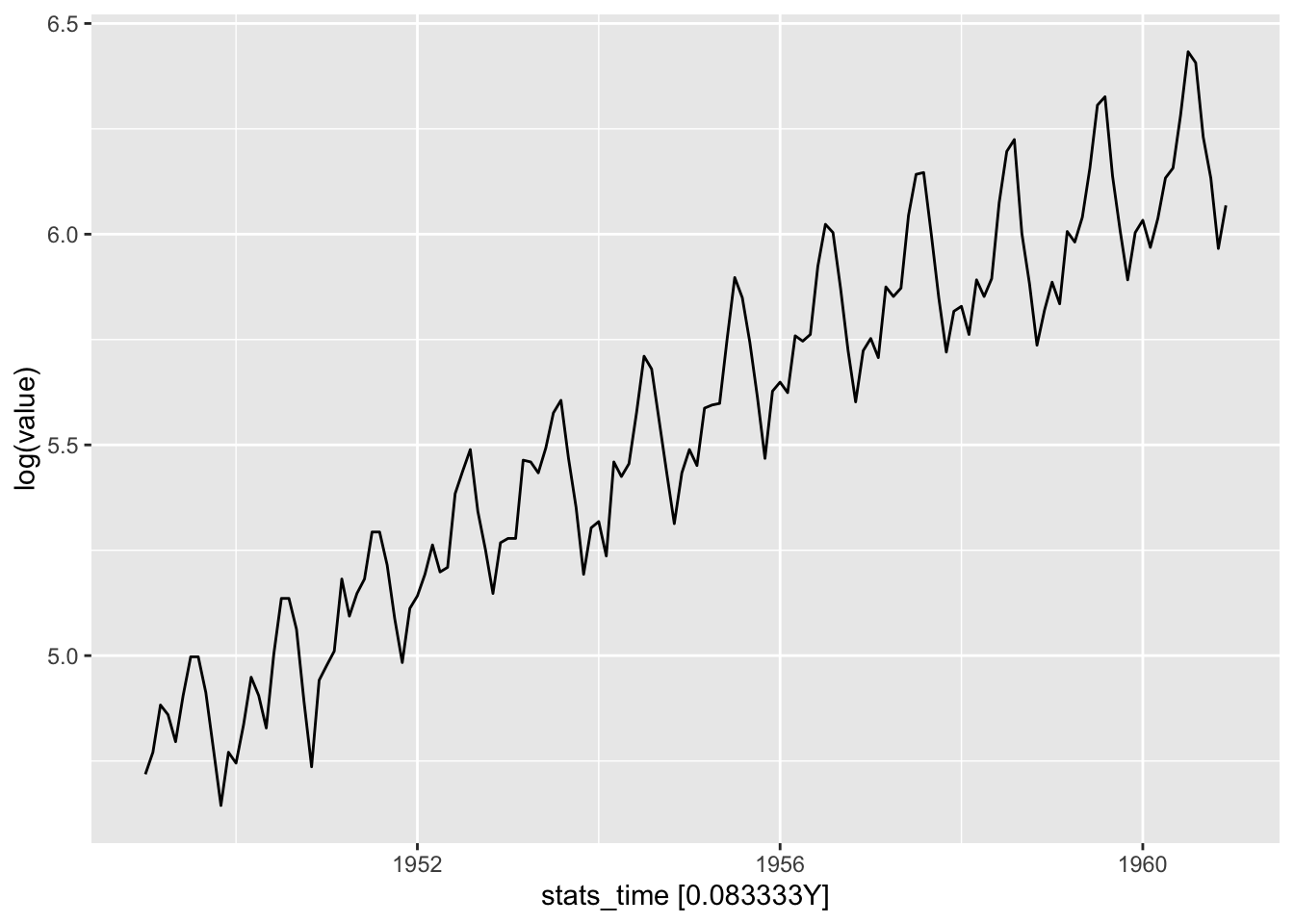
c(mean(time(AP)), sd(time(AP)))[1] 1954.958333 3.476109c(mean(pull(ap, stats_time)),
sd(pull(ap, stats_time)))[1] 1954.958333 3.476109AP.lm1 <- lm(log(AP) ~ TIME + I(TIME^2) + I(TIME^3) +
I(TIME^4) + SIN[,1] + COS[,1] + SIN[,2] + COS[,2] +
SIN[,3] + COS[,3] + SIN[,4] + COS[,4] +
SIN[,5] + COS[,5] + SIN[,6] + COS[,6])
AP.lm2 <- lm(log(AP) ~ TIME + I(TIME^2) +
SIN[,1] + COS[,1] + SIN[,2] + COS[,2] +
SIN[,3] + SIN[,4] + COS[,4] + SIN[,5])
list(lm1 = coef(AP.lm1)/sqrt(diag(vcov(AP.lm1))),
lm2 = coef(AP.lm2)/sqrt(diag(vcov(AP.lm2))))$lm1
(Intercept) TIME I(TIME^2) I(TIME^3) I(TIME^4) SIN[, 1]
741.88611010 42.33701405 -4.15995145 -0.74932492 1.86850936 4.86959429
COS[, 1] SIN[, 2] COS[, 2] SIN[, 3] COS[, 3] SIN[, 4]
-25.43917945 10.39544938 9.83043452 -4.84196512 -1.51378621 -5.66320251
COS[, 4] SIN[, 5] COS[, 5] SIN[, 6] COS[, 6]
1.91354935 -3.76145265 1.01896758 0.07897041 -0.47003062
$lm2
(Intercept) TIME I(TIME^2) SIN[, 1] COS[, 1] SIN[, 2]
922.625445 103.520628 -8.235569 4.916041 -25.813114 10.355319
COS[, 2] SIN[, 3] SIN[, 4] COS[, 4] SIN[, 5]
9.961335 -4.789997 -5.612235 1.949391 -3.730666 ap_lm <- ap |>
model(
lm1 = TSLM(log(value) ~ TIME +
I(TIME^2) + I(TIME^3) + I(TIME^4) +
sin1 + cos1 + sin2 + cos2 + sin3 + cos3 +
sin4 + cos4 + sin5 + cos5 + sin6 + cos6),
lm2 = TSLM(log(value) ~ TIME + I(TIME^2) + sin1 + cos1 +
sin2 + cos2 + sin3 + sin4 + cos4 + sin5))
tidy(ap_lm) |>
mutate(calc = estimate / std.error) |>
select(.model, term, estimate, std.error, calc) |>
print(n = 30)# A tibble: 28 × 5
.model term estimate std.error calc
<chr> <chr> <dbl> <dbl> <dbl>
1 lm1 (Intercept) 5.59e+0 7.50e-3 745.
2 lm1 TIME 4.27e-1 1.01e-2 42.4
3 lm1 I(TIME^2) -6.55e-2 1.58e-2 -4.14
4 lm1 I(TIME^3) -3.87e-3 5.19e-3 -0.745
5 lm1 I(TIME^4) 1.10e-2 5.95e-3 1.85
6 lm1 sin1 2.78e-2 5.70e-3 4.87
7 lm1 cos1 -1.48e-1 5.67e-3 -26.1
8 lm1 sin2 5.92e-2 6.57e-3 9.01
9 lm1 cos2 5.66e-2 5.66e-3 10.0
10 lm1 sin3 -2.74e-2 5.66e-3 -4.84
11 lm1 cos3 -8.81e-3 5.67e-3 -1.55
12 lm1 sin4 -3.21e-2 5.66e-3 -5.67
13 lm1 cos4 1.10e-2 5.66e-3 1.94
14 lm1 sin5 -2.13e-2 5.67e-3 -3.76
15 lm1 cos5 5.81e-3 5.66e-3 1.03
16 lm1 sin6 -1.02e+8 9.91e+8 -0.103
17 lm1 cos6 -3.26e-3 4.76e-3 -0.684
18 lm2 (Intercept) 5.58e+0 6.05e-3 923.
19 lm2 TIME 4.20e-1 4.06e-3 104.
20 lm2 I(TIME^2) -3.74e-2 4.54e-3 -8.24
21 lm2 sin1 2.81e-2 5.71e-3 4.92
22 lm2 cos1 -1.47e-1 5.70e-3 -25.8
23 lm2 sin2 5.91e-2 5.70e-3 10.4
24 lm2 cos2 5.68e-2 5.70e-3 9.96
25 lm2 sin3 -2.73e-2 5.70e-3 -4.79
26 lm2 sin4 -3.20e-2 5.70e-3 -5.61
27 lm2 cos4 1.11e-2 5.70e-3 1.95
28 lm2 sin5 -2.13e-2 5.70e-3 -3.73 c(AIC(AP.lm1), AIC(AP.lm2))[1] -447.9290 -451.0749glance(ap_lm) |> select(.model, AIC, AICc, BIC)# A tibble: 2 × 4
.model AIC AICc BIC
<chr> <dbl> <dbl> <dbl>
1 lm1 -857. -851. -803.
2 lm2 -860. -857. -824.acf(resid(AP.lm2))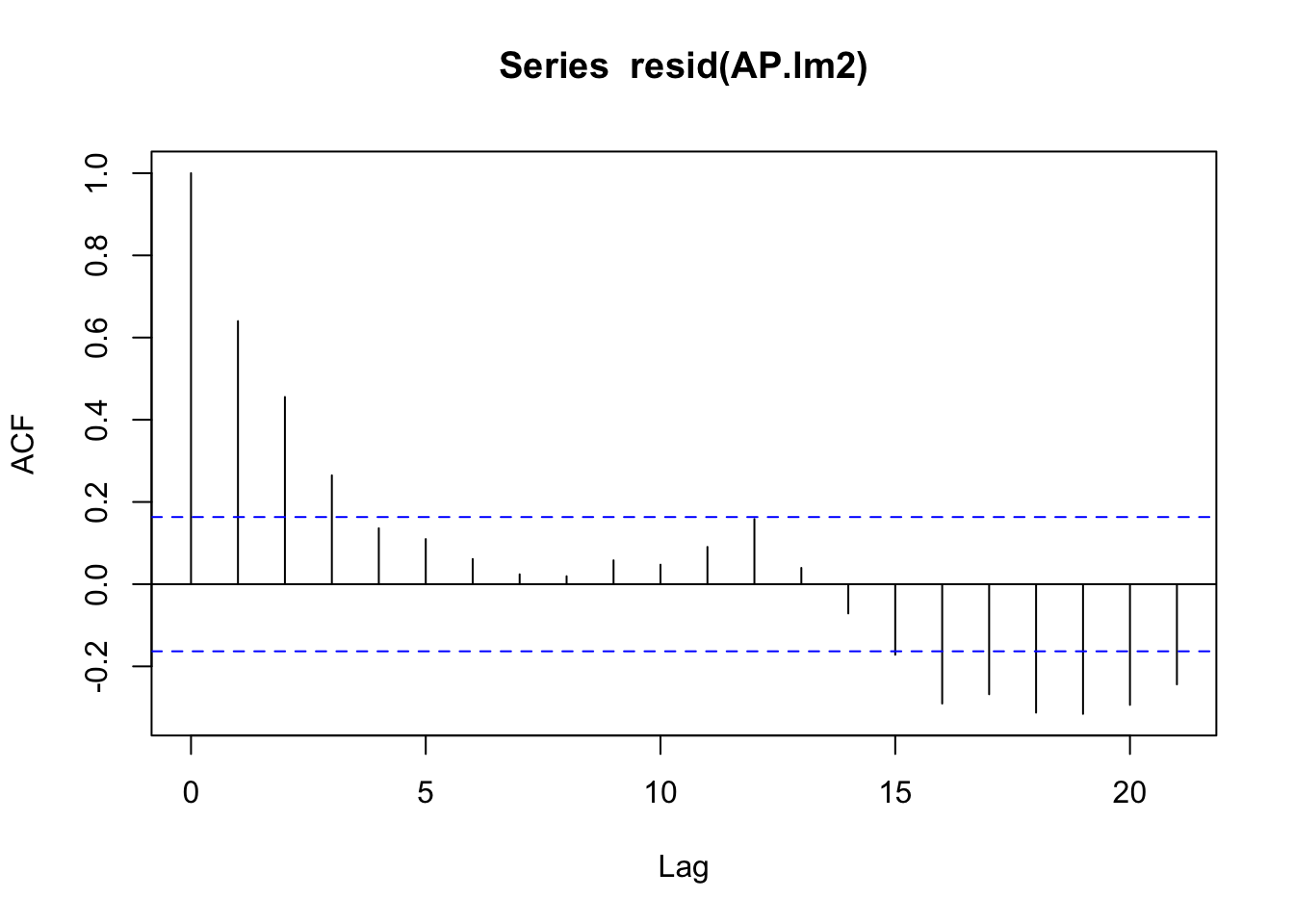
ap_lm |>
select(lm2) |>
residuals() |>
feasts::ACF() |>
autoplot()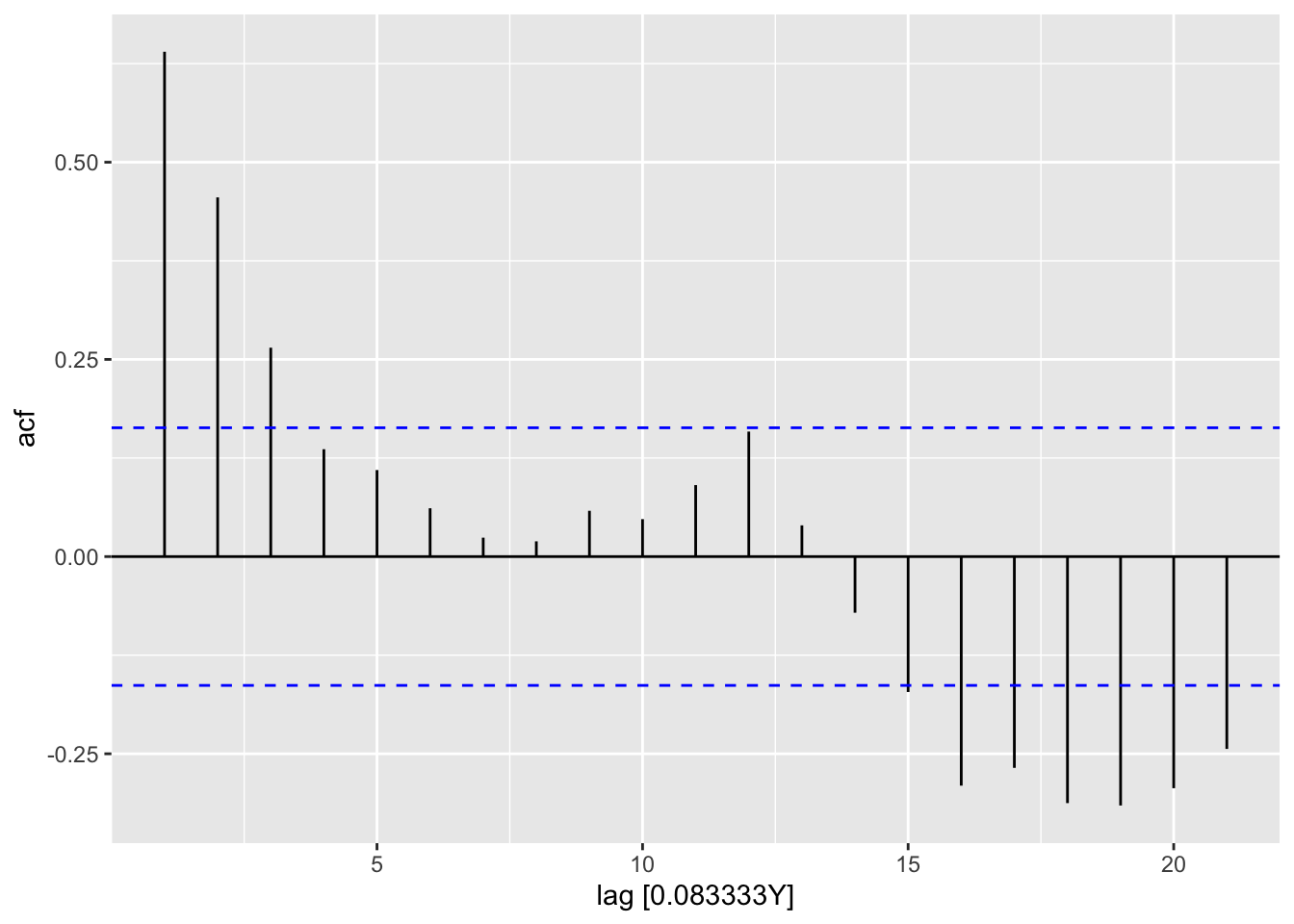
AP.gls <- gls(log(AP) ~ TIME + I(TIME^2) + SIN[,1] + COS[,1] +
SIN[,2] + COS[,2] + SIN[,3] + SIN[,4] + COS[,4] + SIN[,5],
cor = corAR1(0.6))
coef(AP.gls)/sqrt(diag(vcov(AP.gls)))(Intercept) TIME I(TIME^2) SIN[, 1] COS[, 1] SIN[, 2]
398.841575 45.850520 -3.651287 3.299713 -18.177844 11.768934
COS[, 2] SIN[, 3] SIN[, 4] COS[, 4] SIN[, 5]
11.432678 -7.630660 -10.749938 3.571395 -7.920777 ap_gls <- linear_reg() |>
set_engine("gls", correlation = nlme::corAR1(0.6)) |>
fit(log(value) ~ TIME + I(TIME^2) + sin1 + cos1 +
sin2 + cos2 + sin3 + sin4 + cos4 + sin5, data = ap)
broom.mixed::tidy(ap_gls) |>
mutate(calc = estimate / std.error) |>
select(term, estimate, std.error, calc)# A tibble: 11 × 4
term estimate std.error calc
<chr> <dbl> <dbl> <dbl>
1 (Intercept) 5.58 0.0140 399.
2 TIME 0.418 0.00913 45.9
3 I(TIME^2) -0.0364 0.00996 -3.65
4 sin1 0.0270 0.00819 3.30
5 cos1 -0.147 0.00810 -18.2
6 sin2 0.0584 0.00496 11.8
7 cos2 0.0565 0.00494 11.4
8 sin3 -0.0277 0.00363 -7.63
9 sin4 -0.0322 0.00300 -10.7
10 cos4 0.0107 0.00301 3.57
11 sin5 -0.0214 0.00270 -7.92AP.ar <- ar(resid(AP.lm2), order = 1, method = "mle")
AP.ar$ar[1] 0.6410685ap_ar <- augment(ap_gls, ap) |>
as_tsibble(index = stats_time) |>
model(resid = AR(.resid ~ order(1)))
tidy(ap_ar)# A tibble: 2 × 6
.model term estimate std.error statistic p.value
<chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 resid constant 13.5 6.97 1.94 5.50e- 2
2 resid ar1 0.959 0.0234 41.1 1.14e-80acf(AP.ar$res[-1])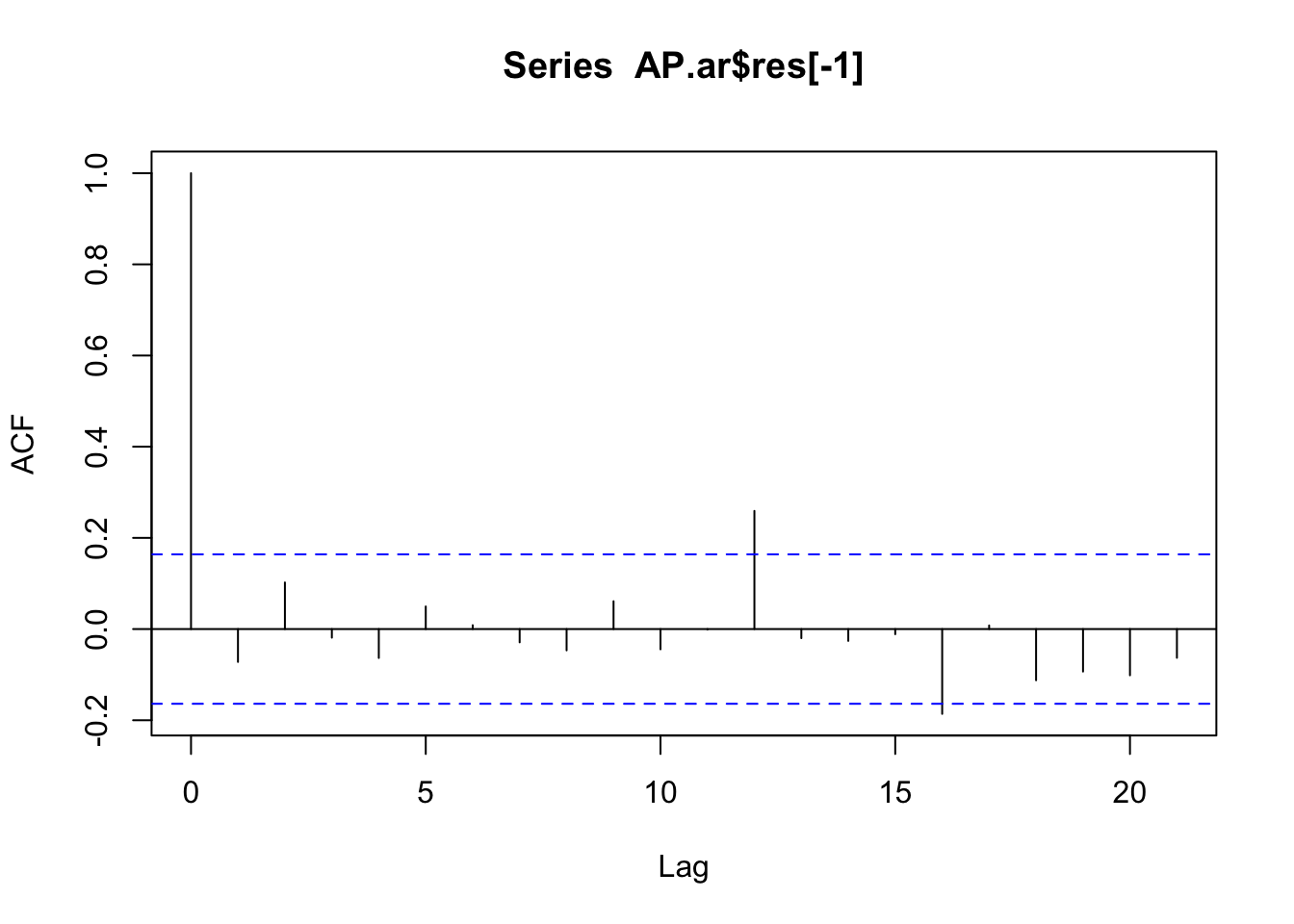
residuals(ap_ar) |>
feasts::ACF(.var = .resid) |>
autoplot()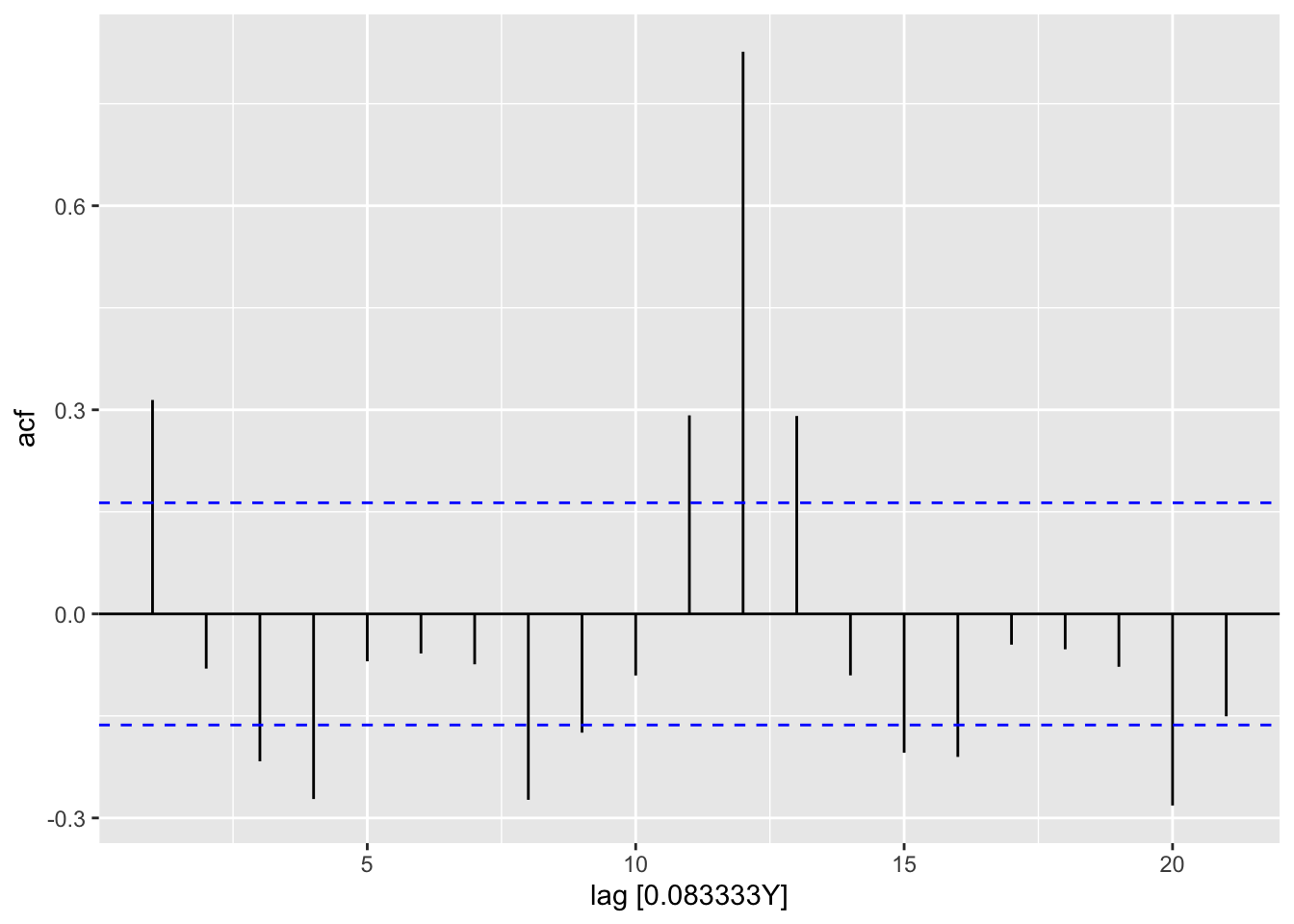
data(AirPassengers)
AP <- AirPassengers
SIN <- COS <- matrix(nr = length(AP), nc = 6)
for (i in 1:6) {
SIN[, i] <- sin(2 * pi * i * time(AP))
COS[, i] <- cos(2 * pi * i * time(AP))
}
TIME <- (time(AP) - mean(time(AP)))/sd(time(AP))
plot(AP)
plot(log(AP))
mean(time(AP))
sd(time(AP))
AP.lm1 <- lm(log(AP) ~ TIME + I(TIME^2) + I(TIME^3) + I(TIME^4) +
SIN[,1] + COS[,1] + SIN[,2] + COS[,2] + SIN[,3] + COS[,3] +
SIN[,4] + COS[,4] + SIN[,5] + COS[,5] + SIN[,6] + COS[,6])
AP.lm2 <- lm(log(AP) ~ TIME + I(TIME^2) + SIN[,1] + COS[,1] +
SIN[,2] + COS[,2] + SIN[,3] + SIN[,4] + COS[,4] + SIN[,5])
list(lm1 = coef(AP.lm1)/sqrt(diag(vcov(AP.lm1))),
lm2 = coef(AP.lm2)/sqrt(diag(vcov(AP.lm2))))
c(AIC(AP.lm1), AIC(AP.lm2))
acf(resid(AP.lm2))
AP.gls <- gls(log(AP) ~ TIME + I(TIME^2) + SIN[,1] + COS[,1] +
SIN[,2] + COS[,2] + SIN[,3] + SIN[,4] + COS[,4] + SIN[,5],
cor = corAR1(0.6))
coef(AP.gls)/sqrt(diag(vcov(AP.gls)))
AP.ar <- ar(resid(AP.lm2), order = 1, method = "mle")
AP.ar$ar
acf(AP.ar$res[-1])data(AirPassengers)
ap <- as_tsibble(AirPassengers) |>
mutate(
Year = year(index),
Month = month(index),
stats_time = Year + (Month - 1)/12,
TIME = (stats_time - mean(stats_time)) / sd(stats_time),
cos1 = cos(2 * pi * 1 * stats_time),
cos2 = cos(2 * pi * 2 * stats_time),
cos3 = cos(2 * pi * 3 * stats_time),
cos4 = cos(2 * pi * 4 * stats_time),
cos5 = cos(2 * pi * 5 * stats_time),
cos6 = cos(2 * pi * 6 * stats_time),
sin1 = sin(2 * pi * 1 * stats_time),
sin2 = sin(2 * pi * 2 * stats_time),
sin3 = sin(2 * pi * 3 * stats_time),
sin4 = sin(2 * pi * 4 * stats_time),
sin5 = sin(2 * pi * 5 * stats_time),
sin6 = sin(2 * pi * 6 * stats_time)) |>
as_tsibble(index = stats_time)
autoplot(ap)
autoplot(ap, .vars = log(value))
mean(pull(ap, stats_time))
sd(pull(ap, stats_time))
ap_lm <- ap |>
model(
lm1 = TSLM(log(value) ~ TIME +
I(TIME^2) + I(TIME^3) + I(TIME^4) +
sin1 + cos1 + sin2 + cos2 + sin3 + cos3 +
sin4 + cos4 + sin5 + cos5 + sin6 + cos6),
lm2 = TSLM(log(value) ~ TIME + I(TIME^2) + sin1 + cos1 +
sin2 + cos2 + sin3 + sin4 + cos4 + sin5))
tidy(ap_lm) |>
mutate(calc = estimate / std.error) |>
print(n = 30)
glance(ap_lm) |> select(.model, AIC, AICc, BIC)
ap_lm |>
select(lm2) |>
residuals() |>
feasts::ACF() |>
autoplot()
ap_gls <- linear_reg() |>
set_engine("gls", correlation = nlme::corAR1(0.6)) |>
fit(log(value) ~ TIME + I(TIME^2) + sin1 + cos1 +
sin2 + cos2 + sin3 + sin4 + cos4 + sin5, data = ap)
broom.mixed::tidy(ap_gls) |>
mutate(calc = estimate / std.error)
ap_ar <- augment(ap_gls, ap) |>
as_tsibble(index = stats_time) |>
model(resid = AR(.resid ~ order(1)))
tidy(ap_ar)
residuals(ap_ar) |>
feasts::ACF(.var = .resid) |>
autoplot()Book code
set.seed(1)
w <- rnorm(100, sd = 10)
z <- rep(0, 100)
for (t in 2:100) z[t] <- 0.7 * z[t - 1] + w[t]
Time <- 1:100
f <- function(x) exp(1 + 0.05 * x)
x <- f(Time) + z
plot(x, type = "l")
abline(0, 0)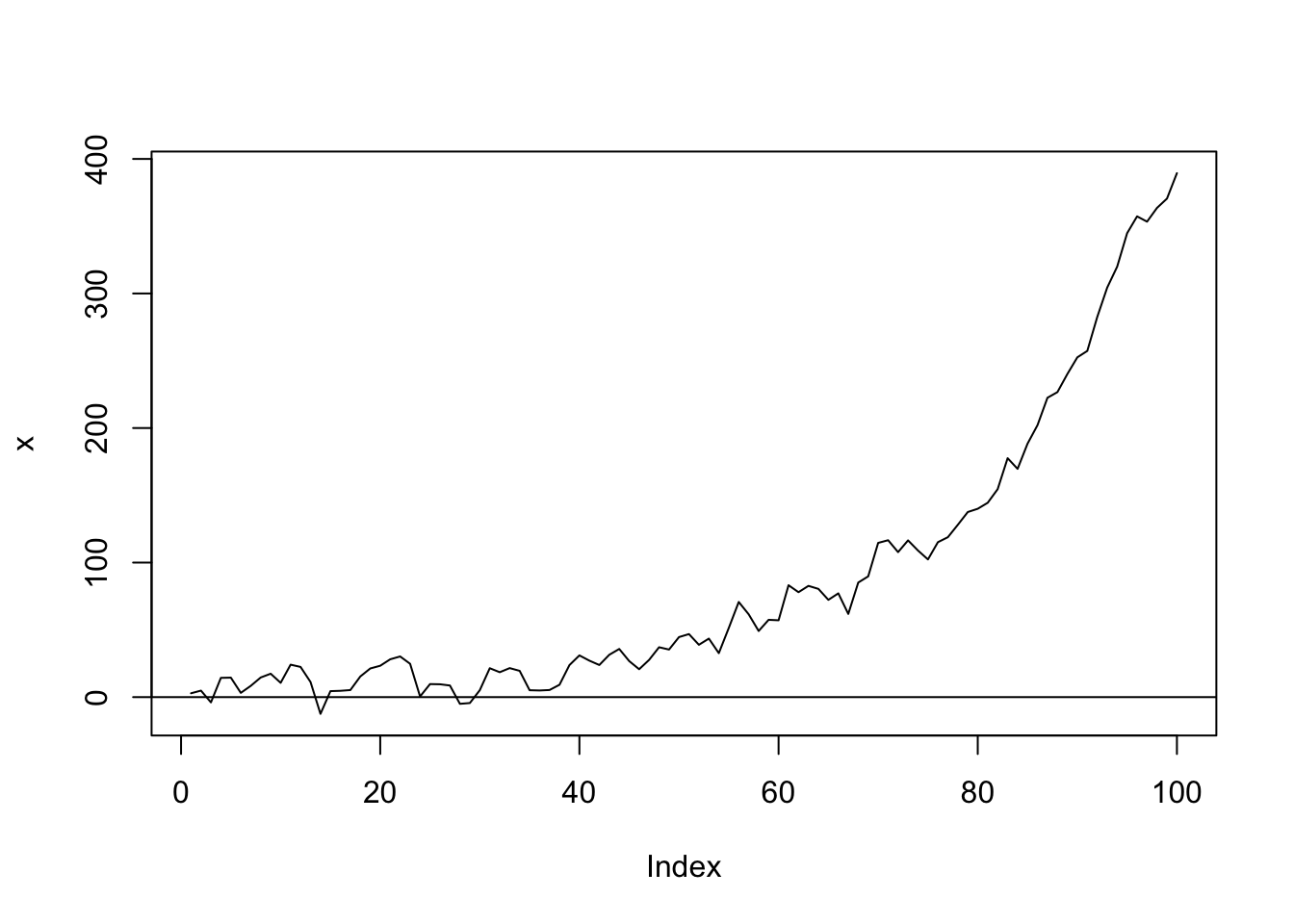
Tidyverts code
set.seed(1)
dat <- tibble(w = rnorm(100, sd = 10)) |>
mutate(
Time = 1:n(),
z = purrr::accumulate2(
lag(w), w,
\(acc, nxt, w) 0.7 * acc + w,
.init = 0)[-1],
x = exp(1 + 0.05 * Time) + z) |>
tsibble::as_tsibble(index = Time)
autoplot(dat, .var = x) +
geom_hline(yintercept = 0)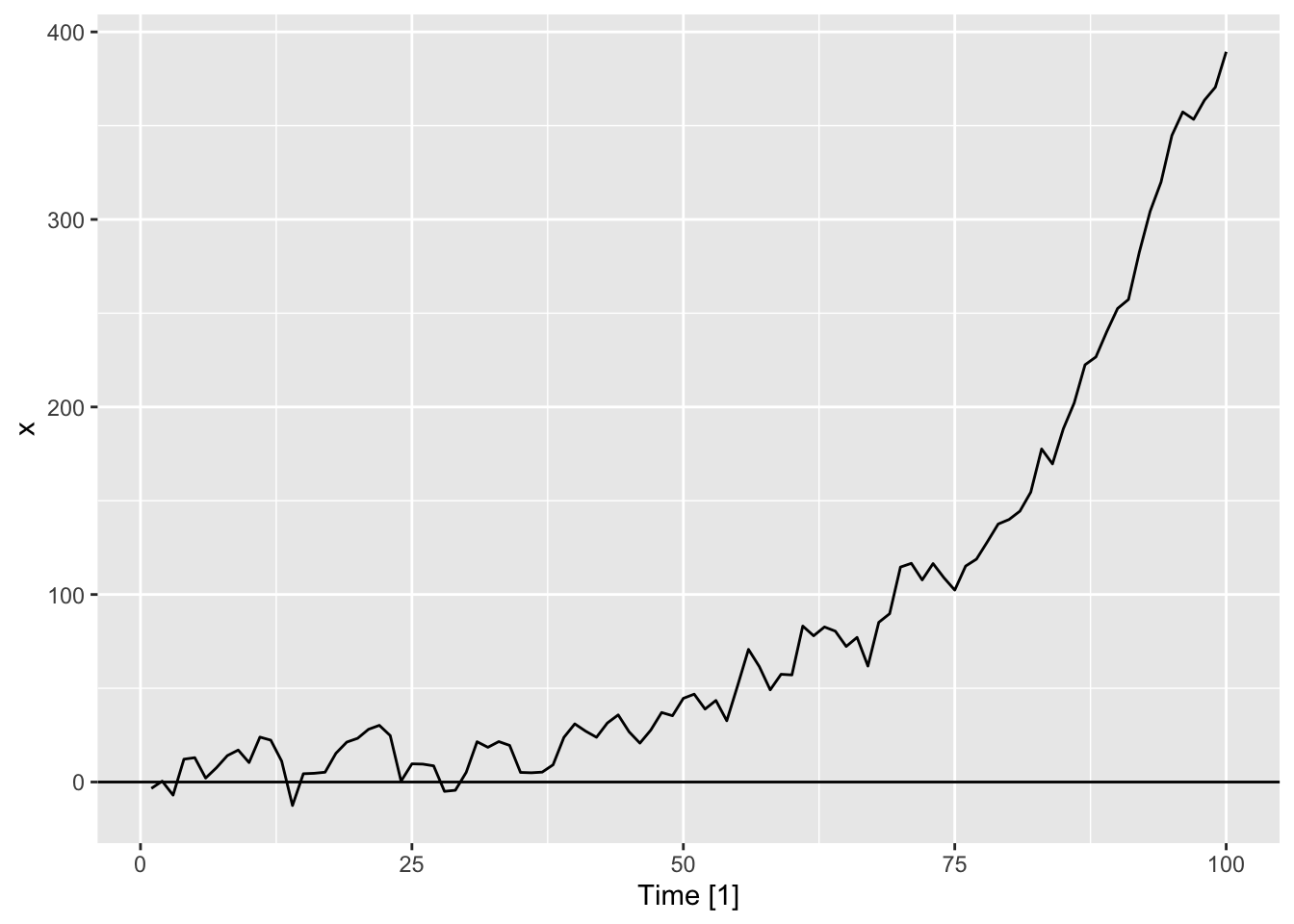
x.nls <- nls(x ~ exp(alp0 + alp1 * Time), start = list(alp0 = 0.1,
alp1 = 0.5))
summary(x.nls)$parameters Estimate Std. Error t value Pr(>|t|)
alp0 1.17640121 0.0742954459 15.83410 9.204061e-29
alp1 0.04828424 0.0008188462 58.96619 2.347546e-78equation <- x ~ exp(alp0 + alp1 * Time)
dat_nls <- nls(equation, data = dat,
start = list(alp0 = 0.1, alp1 = 0.5))
summary(dat_nls)$parameters Estimate Std. Error t value Pr(>|t|)
alp0 1.17299000 0.0743864847 15.76886 1.232132e-28
alp1 0.04832158 0.0008197862 58.94413 2.432829e-78set.seed(1)
w <- rnorm(100, sd = 10)
z <- rep(0, 100)
for (t in 2:100) z[t] <- 0.7 * z[t - 1] + w[t]
Time <- 1:100
f <- function(x) exp(1 + 0.05 * x)
x <- f(Time) + z
plot(x, type = "l")
abline(0, 0)
x.nls <- nls(x ~ exp(alp0 + alp1 * Time), start = list(alp0 = 0.1,
alp1 = 0.5))
summary(x.nls)$parametersset.seed(1)
dat <- tibble(w = rnorm(100, sd = 10)) |>
mutate(
Time = 1:n(),
z = purrr::accumulate2(
lag(w), w,
\(acc, nxt, w) 0.7 * acc + w,
.init = 0)[-1],
x = exp(1 + 0.05 * Time) + z) |>
tsibble::as_tsibble(index = Time)
autoplot(dat, .var = x) +
geom_hline(yintercept = 0)
equation <- x ~ exp(alp0 + alp1 * Time)
dat_nls <- nls(equation, data = dat,
start = list(alp0 = 0.1, alp1 = 0.5))
summary(dat_nls)$parametersBook code
new.t <- time(ts(start = 1961, end = c(1970, 12), fr = 12))
TIME <- (new.t - mean(time(AP)))/sd(time(AP))
SIN <- COS <- matrix(nr = length(new.t), nc = 6)
for (i in 1:6) {
COS[, i] <- cos(2 * pi * i * new.t)
SIN[, i] <- sin(2 * pi * i * new.t)
}
SIN <- SIN[, -6]
new.dat <- data.frame(TIME = as.vector(TIME), SIN = SIN,
COS = COS)
AP.pred.ts <- exp(ts(predict(AP.lm2, new.dat), st = 1961,
fr = 12))
ts.plot(log(AP), log(AP.pred.ts), lty = 1:2)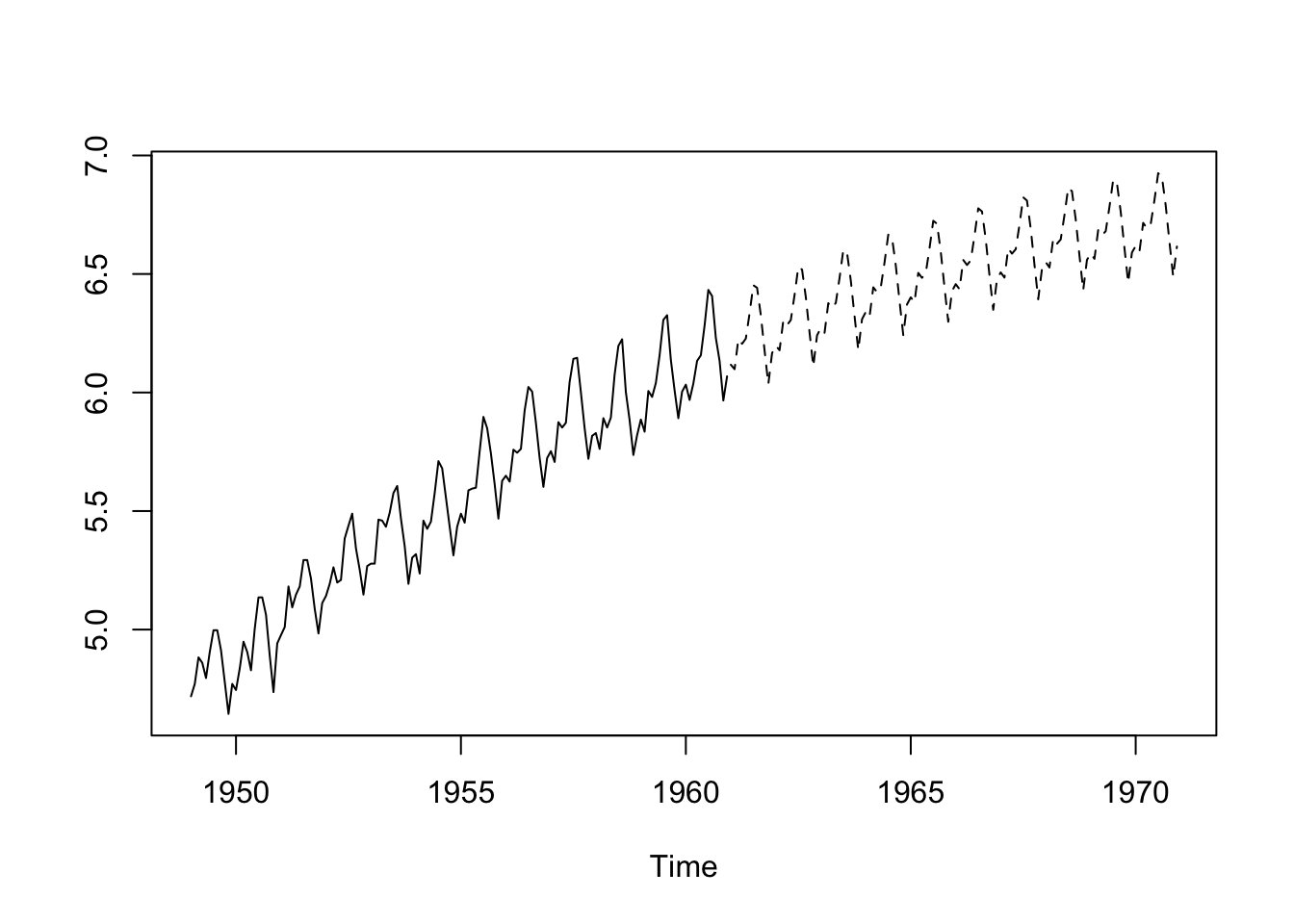
Tidyverts code
new_dat <- tibble(new_t = seq(1961, 1970.99, 1/12)) |>
mutate(
TIME = (new_t - mean(pull(ap, stats_time))) / sd(pull(ap, stats_time)),
cos1 = cos(2 * pi * 1 * new_t),
cos2 = cos(2 * pi * 2 * new_t),
cos3 = cos(2 * pi * 3 * new_t),
cos4 = cos(2 * pi * 4 * new_t),
cos5 = cos(2 * pi * 5 * new_t),
cos6 = cos(2 * pi * 6 * new_t),
sin1 = sin(2 * pi * 1 * new_t),
sin2 = sin(2 * pi * 2 * new_t),
sin3 = sin(2 * pi * 3 * new_t),
sin4 = sin(2 * pi * 4 * new_t),
sin5 = sin(2 * pi * 5 * new_t)) |>
rename(stats_time = new_t) |>
as_tsibble(index = stats_time)
ap_lm |>
select(lm2) |>
forecast(new_data = new_dat) |>
autoplot(ap, level = 95) +
scale_y_continuous(trans = "log", labels = trans_format("log"))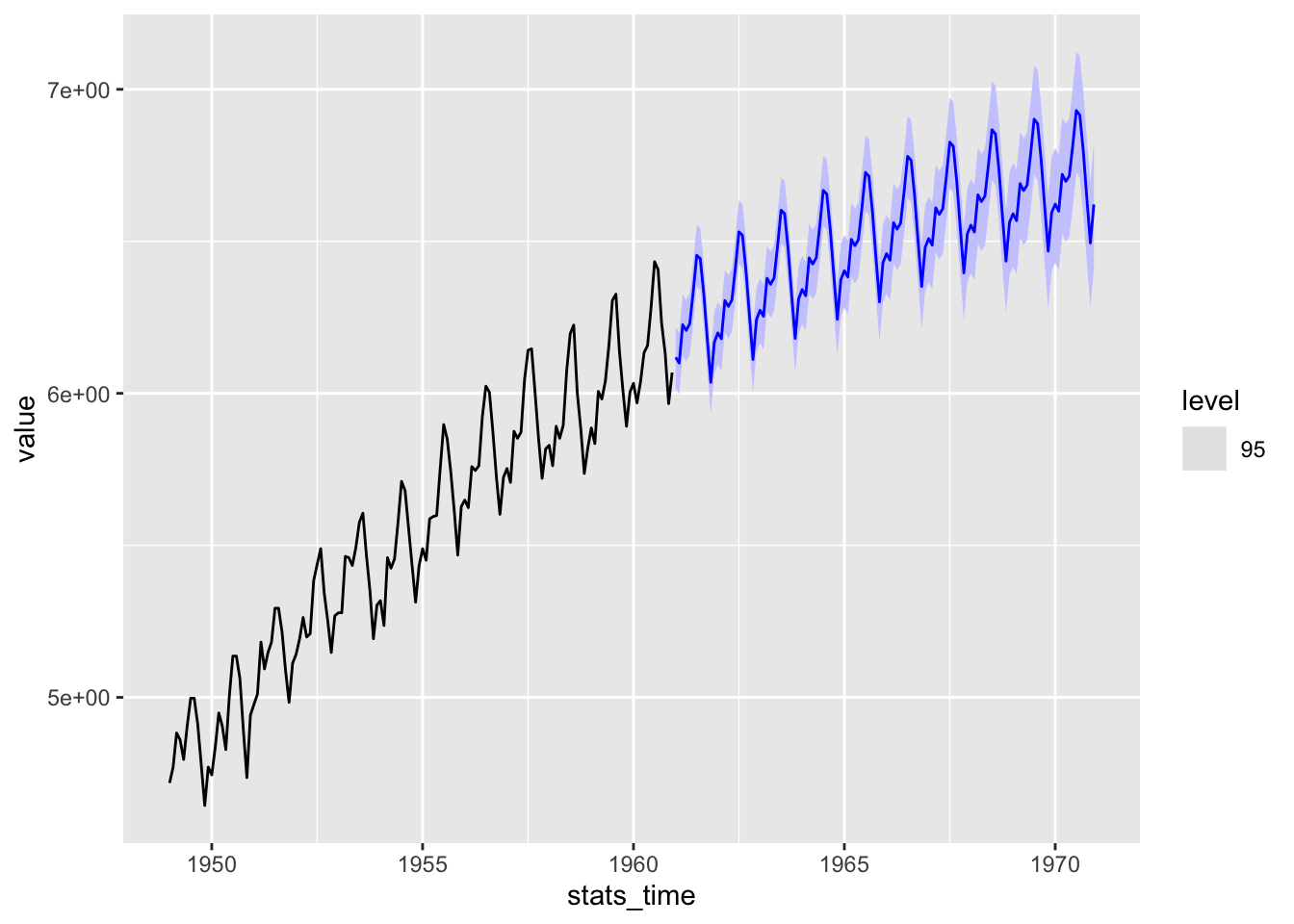
ts.plot(AP, AP.pred.ts, lty = 1:2)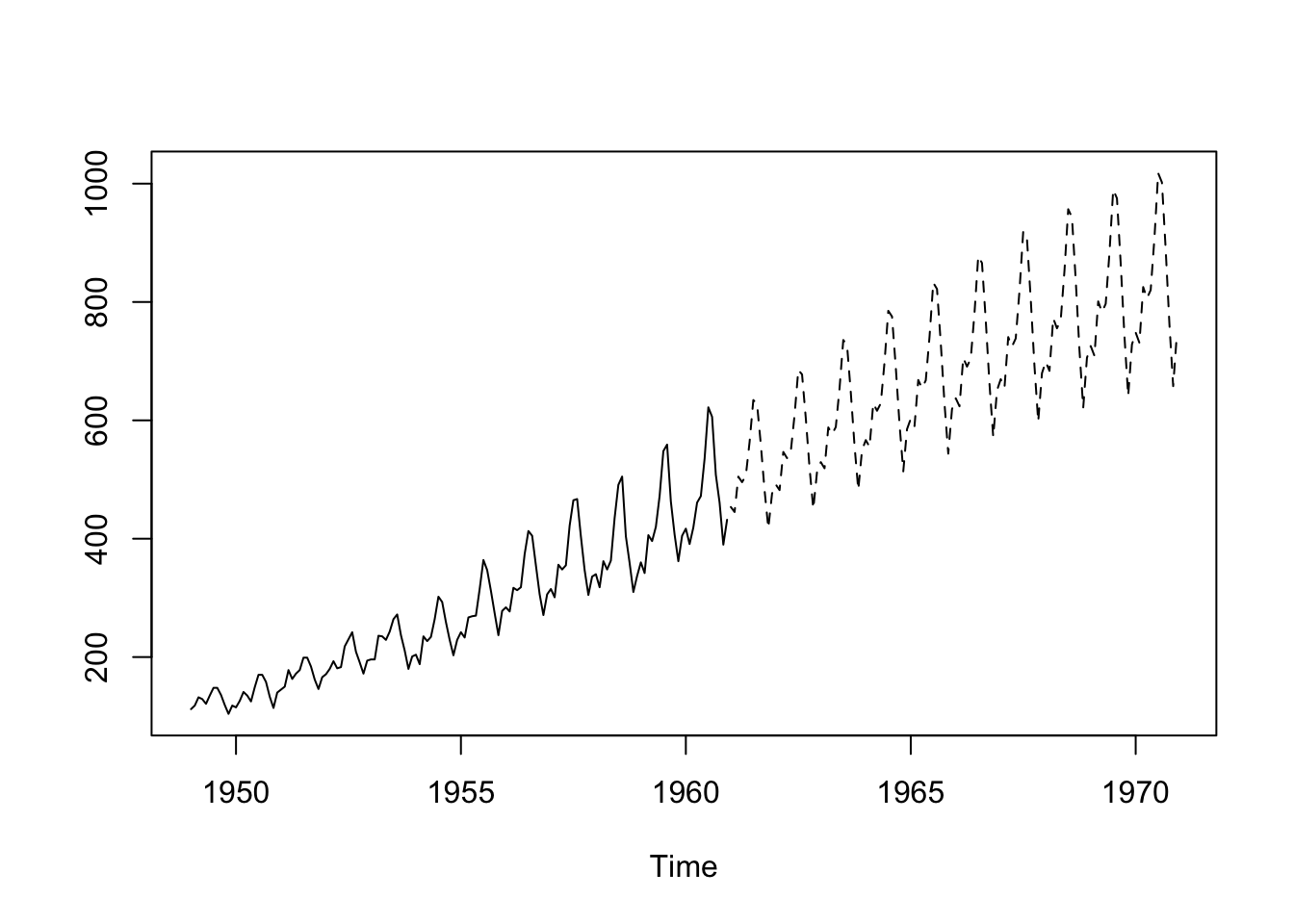
ap_lm |>
select(lm2) |>
forecast(new_data = new_dat) |>
autoplot(ap, level = 95)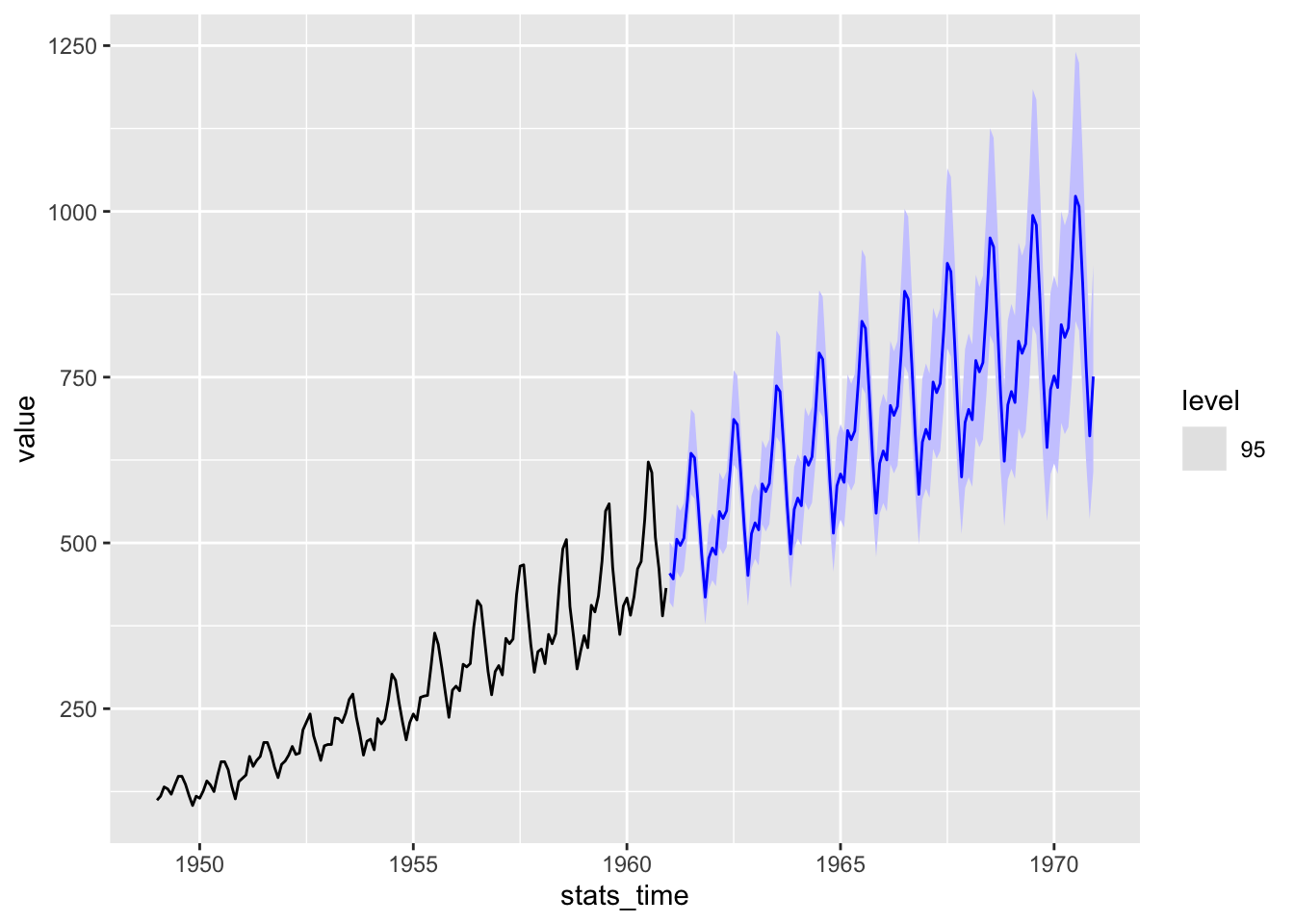
new.t <- time(ts(start = 1961, end = c(1970, 12), fr = 12))
TIME <- (new.t - mean(time(AP)))/sd(time(AP))
SIN <- COS <- matrix(nr = length(new.t), nc = 6)
for (i in 1:6) {
COS[, i] <- cos(2 * pi * i * new.t)
SIN[, i] <- sin(2 * pi * i * new.t)
}
SIN <- SIN[, -6]
new.dat <- data.frame(TIME = as.vector(TIME), SIN = SIN,
COS = COS)
AP.pred.ts <- exp(ts(predict(AP.lm2, new.dat), st = 1961,
fr = 12))
ts.plot(log(AP), log(AP.pred.ts), lty = 1:2)
ts.plot(AP, AP.pred.ts, lty = 1:2)new_dat <- tibble(new_t = seq(1961, 1970.99, 1/12)) |>
mutate(
TIME = (new_t - mean(pull(ap, stats_time))) / sd(pull(ap, stats_time)),
cos1 = cos(2 * pi * 1 * new_t),
cos2 = cos(2 * pi * 2 * new_t),
cos3 = cos(2 * pi * 3 * new_t),
cos4 = cos(2 * pi * 4 * new_t),
cos5 = cos(2 * pi * 5 * new_t),
cos6 = cos(2 * pi * 6 * new_t),
sin1 = sin(2 * pi * 1 * new_t),
sin2 = sin(2 * pi * 2 * new_t),
sin3 = sin(2 * pi * 3 * new_t),
sin4 = sin(2 * pi * 4 * new_t),
sin5 = sin(2 * pi * 5 * new_t)) |>
rename(stats_time = new_t) |>
as_tsibble(index = stats_time)
ap_lm |>
select(lm2) |>
forecast(new_data = new_dat) |>
autoplot(ap, level = 95) +
scale_y_continuous(trans = "log", labels = trans_format("log"))
ap_lm |>
select(lm2) |>
forecast(new_data = new_dat) |>
autoplot(ap, level = 95)Book code
set.seed(1)
sigma <- 1
w <- rnorm(1e+06, sd = sigma)
c(mean(w), mean(exp(w)), exp(sigma^2/2))[1] 4.690776e-05 1.648610e+00 1.648721e+00Tidyverts code
set.seed(1)
sigma <- 1
w <- rnorm(1e+06, sd = sigma)
c(mean(w), mean(exp(w)), exp(sigma^2/2))[1] 4.690776e-05 1.648610e+00 1.648721e+00Book code
summary(AP.lm2)$r.sq[1] 0.9888318Tidyverts code
model_values <- ap_lm |>
select(lm2) |>
glance()
model_values |>
select(r_squared)# A tibble: 1 × 1
r_squared
<dbl>
1 0.989sigma <- summary(AP.lm2)$sigma
lognorm.correction.factor <- exp((1/2) * sigma^2)
empirical.correction.factor <- mean(exp(resid(AP.lm2)))
c(lognorm.correction.factor, empirical.correction.factor)[1] 1.001171 1.001080old <- options(pillar.sigfig = 7)
model_check <- model_values |>
mutate(
sigma = sqrt(sigma2),
lognorm_cf = exp((1/2) * sigma2),
empirical_cf = ap_lm |>
select(lm2) |>
residuals() |>
pull(.resid) |>
exp() |>
mean()) |>
select(.model, r_squared, sigma2, sigma, lognorm_cf, empirical_cf)
model_check# A tibble: 1 × 6
.model r_squared sigma2 sigma lognorm_cf empirical_cf
<chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 lm2 0.9888318 0.002340141 0.04837501 1.001171 1.001080AP.pred.ts <- AP.pred.ts * empirical.correction.factoroptions(old)
ap_pred <- ap_lm |>
select(lm2) |>
forecast(new_data = new_dat) |>
mutate(.mean_correction = .mean * model_check$empirical_cf) |>
select(stats_time, value, .mean, .mean_correction)model_values <- ap_lm |>
select(lm2) |>
glance()
model_values |>
select(r_squared)
old <- options(pillar.sigfig = 7)
model_check <- model_values |>
mutate(
sigma = sqrt(sigma2),
lognorm_cf = exp((1/2) * sigma2),
empirical_cf = ap_lm |>
select(lm2) |>
residuals() |>
pull(.resid) |>
exp() |>
mean()) |>
select(.model, r_squared, sigma2, sigma, lognorm_cf, empirical_cf)
model_check
options(old)
ap_pred <- ap_lm |>
select(lm2) |>
forecast(new_data = new_dat) |>
mutate(.mean_correction = .mean * model_check$empirical_cf) |>
select(stats_time, value, .mean, .mean_correction)R commands used in examplesfable::TSLM(): Fit a linear model with time series componentsparsnip::linear_reg(): Linear regressionparsnip::set_engine(): Declare a computational engine and specific argumentsnlme::corAR1: AR(1) Correlation Structurebroom.mixed::tidy(): Turn an object into a tidy tibblebroom::glance(): Glance at a(n) lm objectggplot2::geom_hline(): Reference lines: horizontal, vertical, and diagonalggplot2::scale_y_continuous(): Position scales for continuous data (x & y)scales::trans_format(): Format labels after transformation