We also need to estimate correlations if we are to generate realistic time series for simulations. The correlation structure of a time series model is defined by the correlation function, and we estimate this from the observed time series.
2.2.1 ‘Expected value’ Modern Look
Book code
<- read.table ("data/Herald.dat" , header = T)# attach (Herald.dat) # This is bad <- Herald.dat # abbreviating longer name. Not in book sum ((HD$ CO - mean (HD$ CO)) * (HD$ Benzoa - mean (HD$ Benzoa))) / (nrow (HD) - 1 )mean ((HD$ CO - mean (HD$ CO)) * (HD$ Benzoa - mean (HD$ Benzoa)))
Tidyverts code
# install.packages("pacman") :: p_load ("tsibble" , "fable" , "feasts" ,"tsibbledata" , "fable.prophet" , "tidyverse" , "patchwork" )<- readr:: read_table ("data/Herald.dat" ) |> rename_all ("str_to_lower" )<- mutate (hd,mc = mean (co), mb = mean (benzoa),equation_i = (co - mc) * (benzoa - mb)summarise (hd_calc, method_1 = sum (equation_i) / (n ()- 1 ),method_2 = mean (equation_i),method_3 = cov (co, benzoa))
# A tibble: 1 × 3
method_1 method_2 method_3
<dbl> <dbl> <dbl>
1 5.51 5.17 5.51
cov (HD$ CO, HD$ Benzoa) / (sd (HD$ CO) * sd (HD$ Benzoa))
summarise (hd_calc, correlation_m1 = cov (co, benzoa) / (sd (co) * sd (benzoa)),correlation_m2 = cor (co, benzoa))
# A tibble: 1 × 2
correlation_m1 correlation_m2
<dbl> <dbl>
1 0.355 0.355
Show the Book code in full
<- read.table ("data/Herald.dat" , header = T)# attach (Herald.dat) # This is bad <- Herald.dat # abbreviating longer name. Not in book sum ((HD$ CO - mean (HD$ CO)) * (HD$ Benzoa - mean (HD$ Benzoa))) / (nrow (HD) - 1 )mean ((HD$ CO - mean (HD$ CO)) * (HD$ Benzoa - mean (HD$ Benzoa)))cov (HD$ CO, HD$ Benzoa)cov (HD$ CO, HD$ Benzoa) / (sd (HD$ CO) * sd (HD$ Benzoa))cor (HD$ CO, HD$ Benzoa)
Show the Tidyverts code in full
# install.packages("pacman") :: p_load ("tsibble" , "fable" , "feasts" ,"tsibbledata" , "fable.prophet" , "tidyverse" )<- readr:: read_table ("data/Herald.dat" ) |> rename_all ("str_to_lower" )<- mutate (hd,mc = mean (co), mb = mean (benzoa),equation_i = (co - mc) * (benzoa - mb)summarise (hd_calc, method_1 = sum (equation_i) / (n ()- 1 ),method_2 = mean (equation_i),method_3 = cov (co, benzoa))summarise (hd_calc, correlation_m1 = cov (co, benzoa) / (sd (co) * sd (benzoa)),correlation_m2 = cor (co, benzoa))
2.25 ‘Autocorrelation’ and ‘Wave heights’ Modern Look
Book code
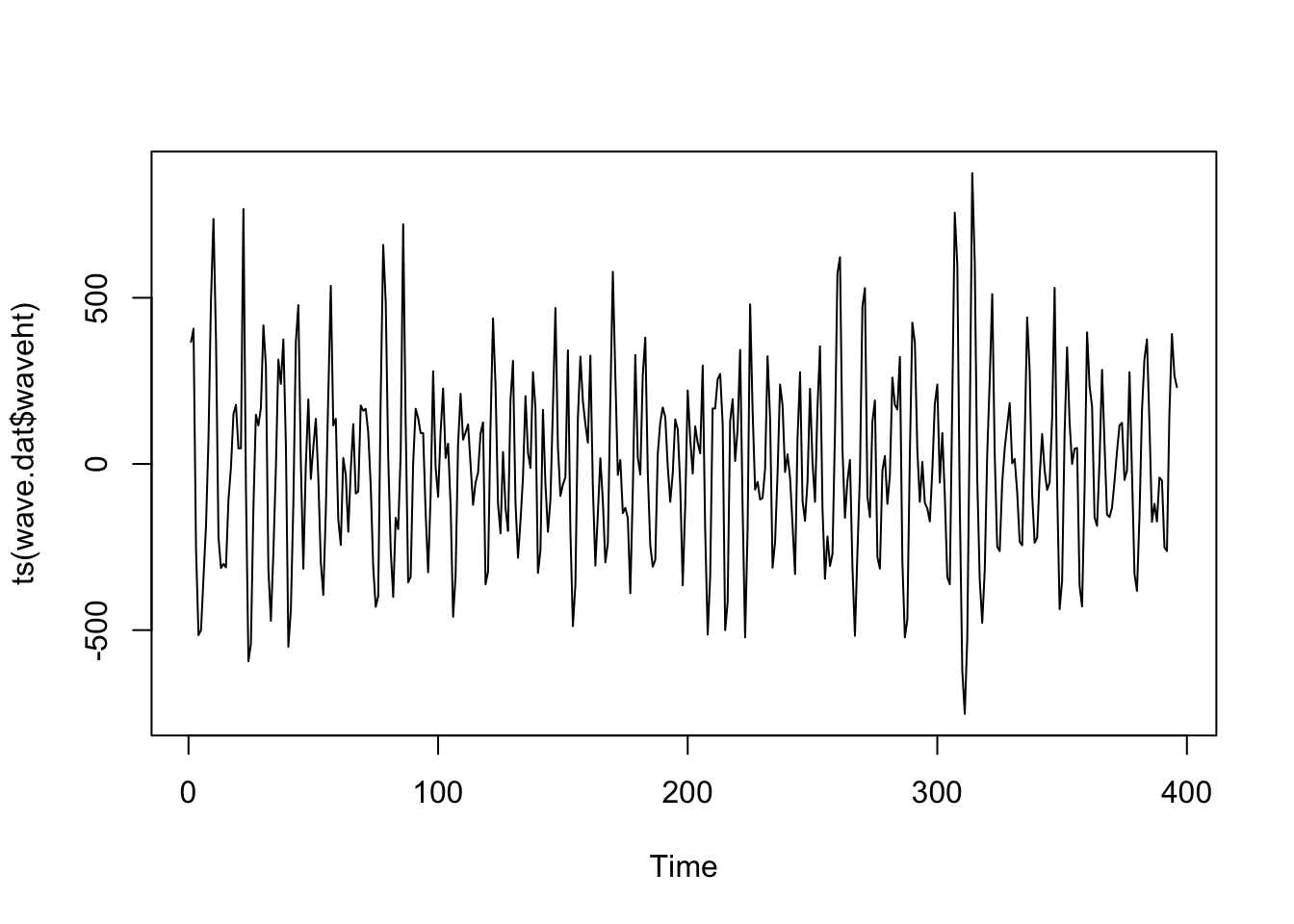
<- read.table ("data/wave.dat" , header= T)# attach(wave.dat) #bf plot (ts (wave.dat$ waveht))
Tidyverts code
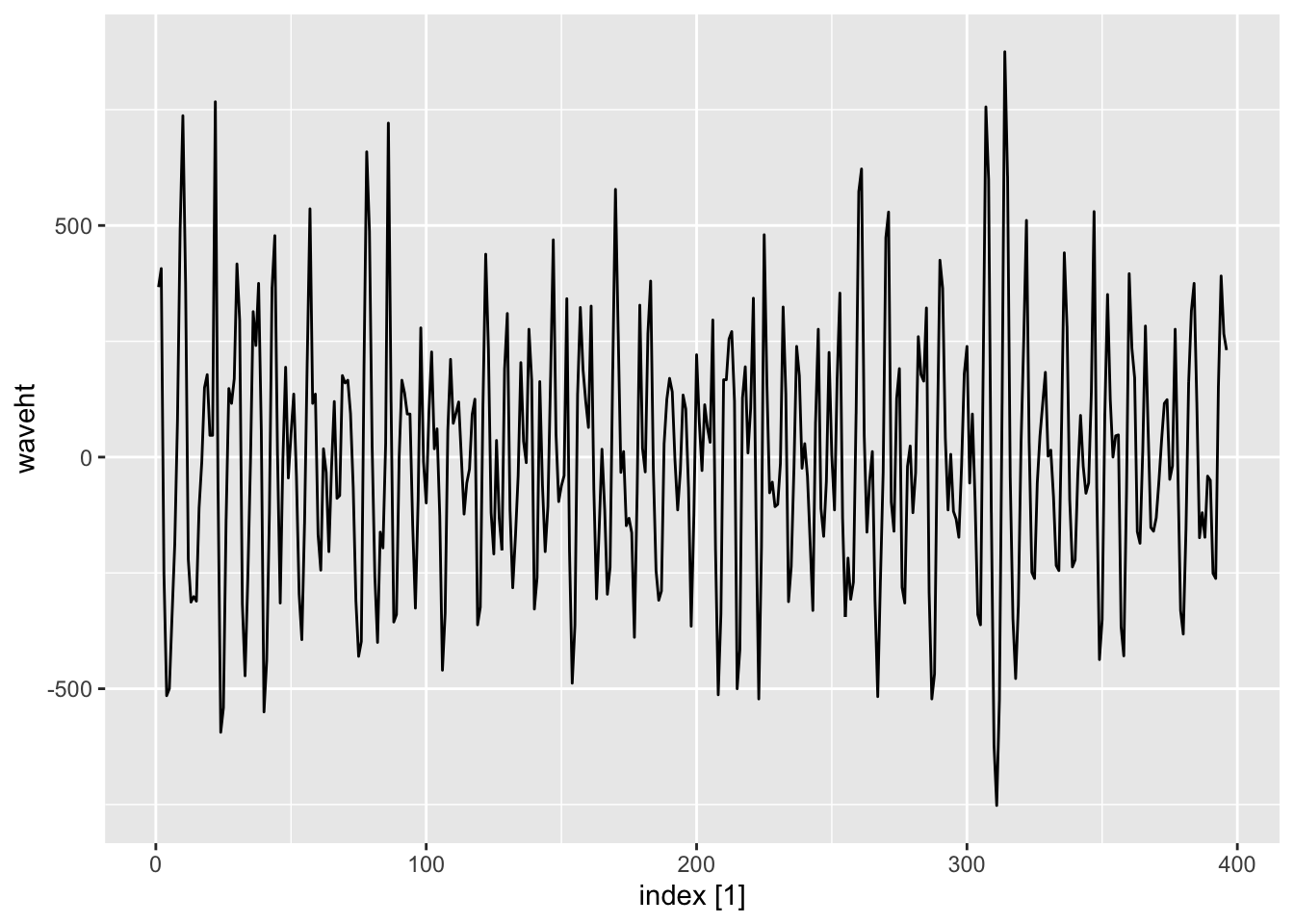
<- read_table ("data/wave.dat" ) |> mutate (index = 1 : n ()) |> as_tsibble (index = index)|> autoplot ()
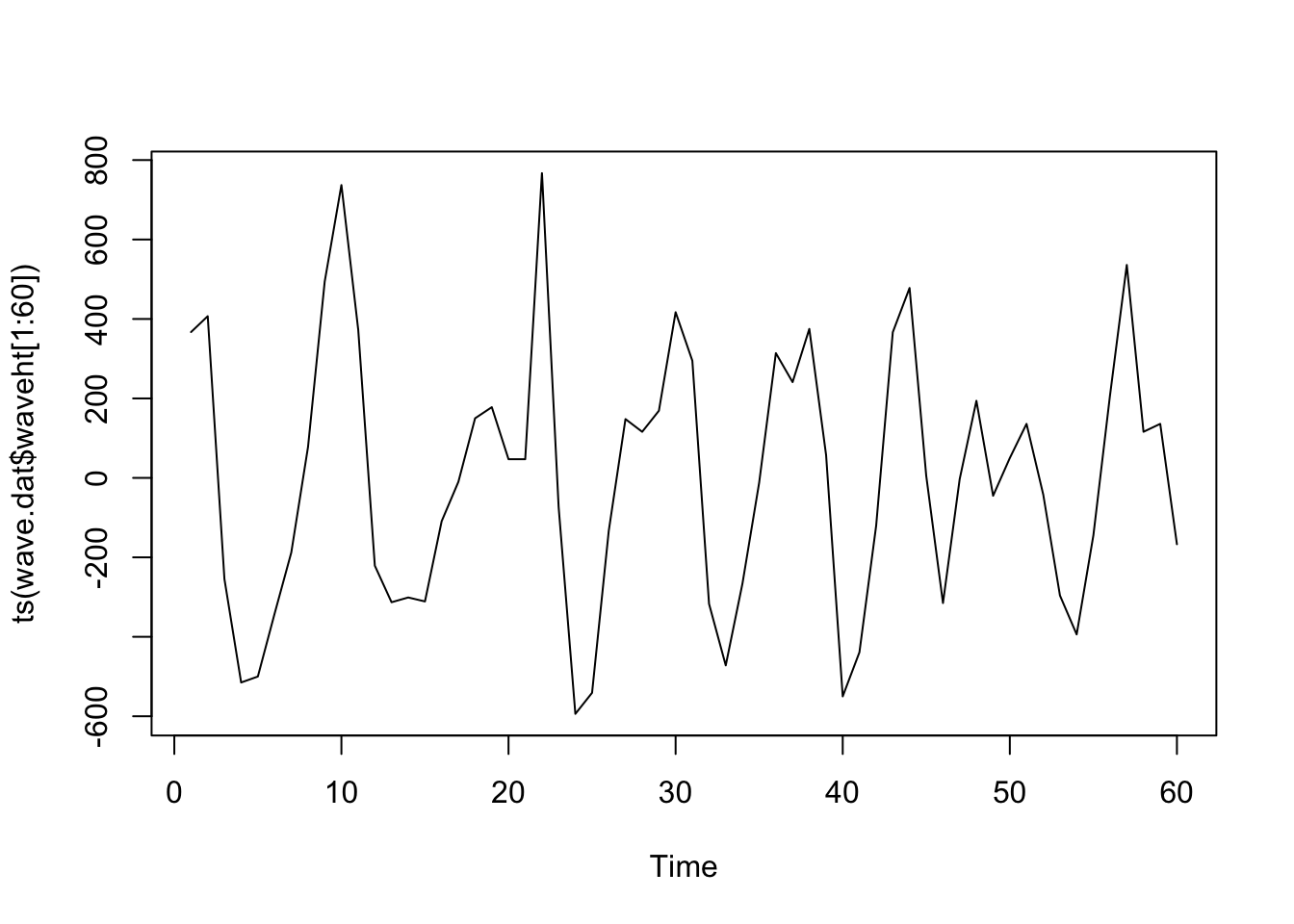
plot (ts (wave.dat$ waveht[1 : 60 ]))
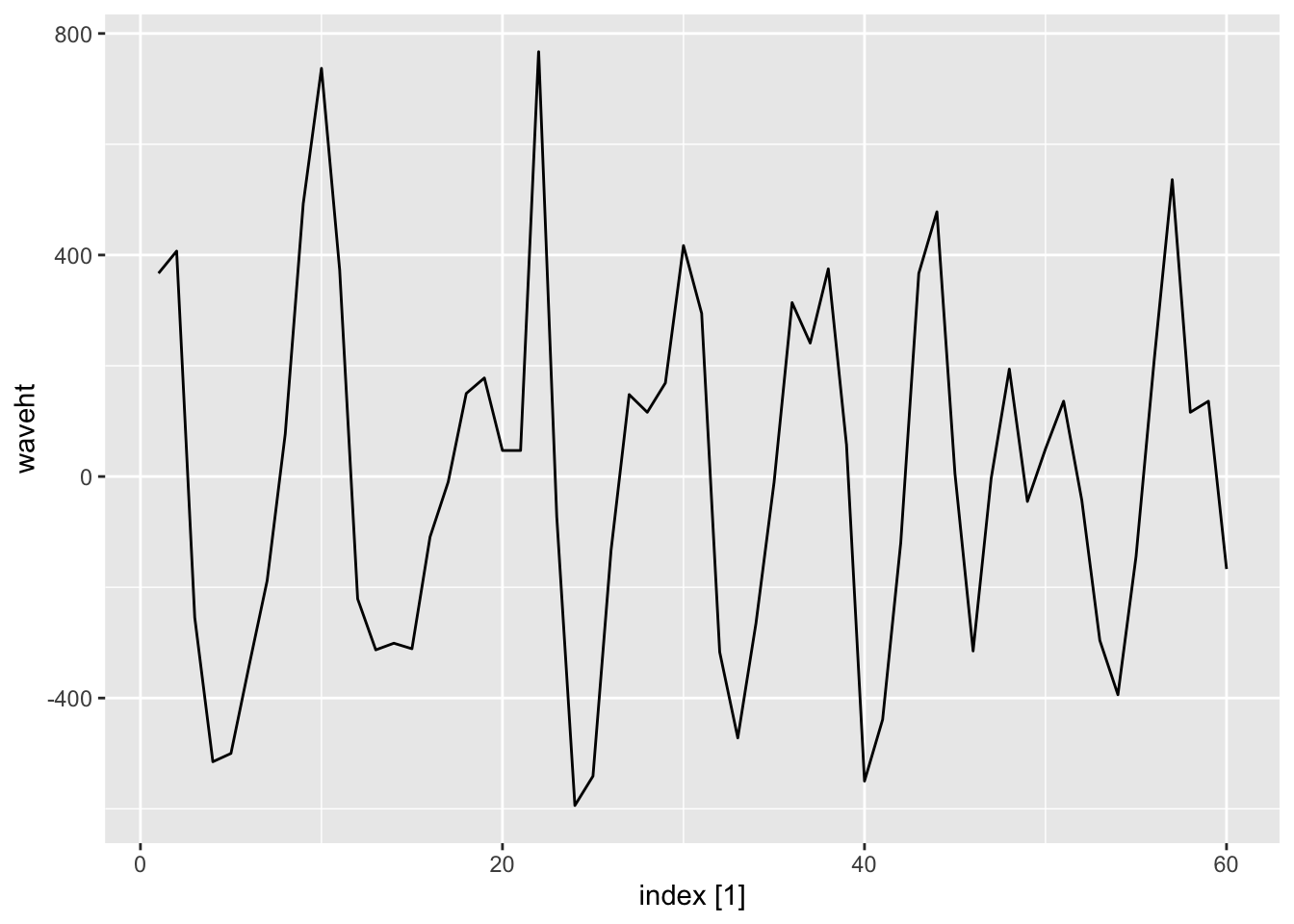
|> slice (1 : 60 ) |> autoplot ()
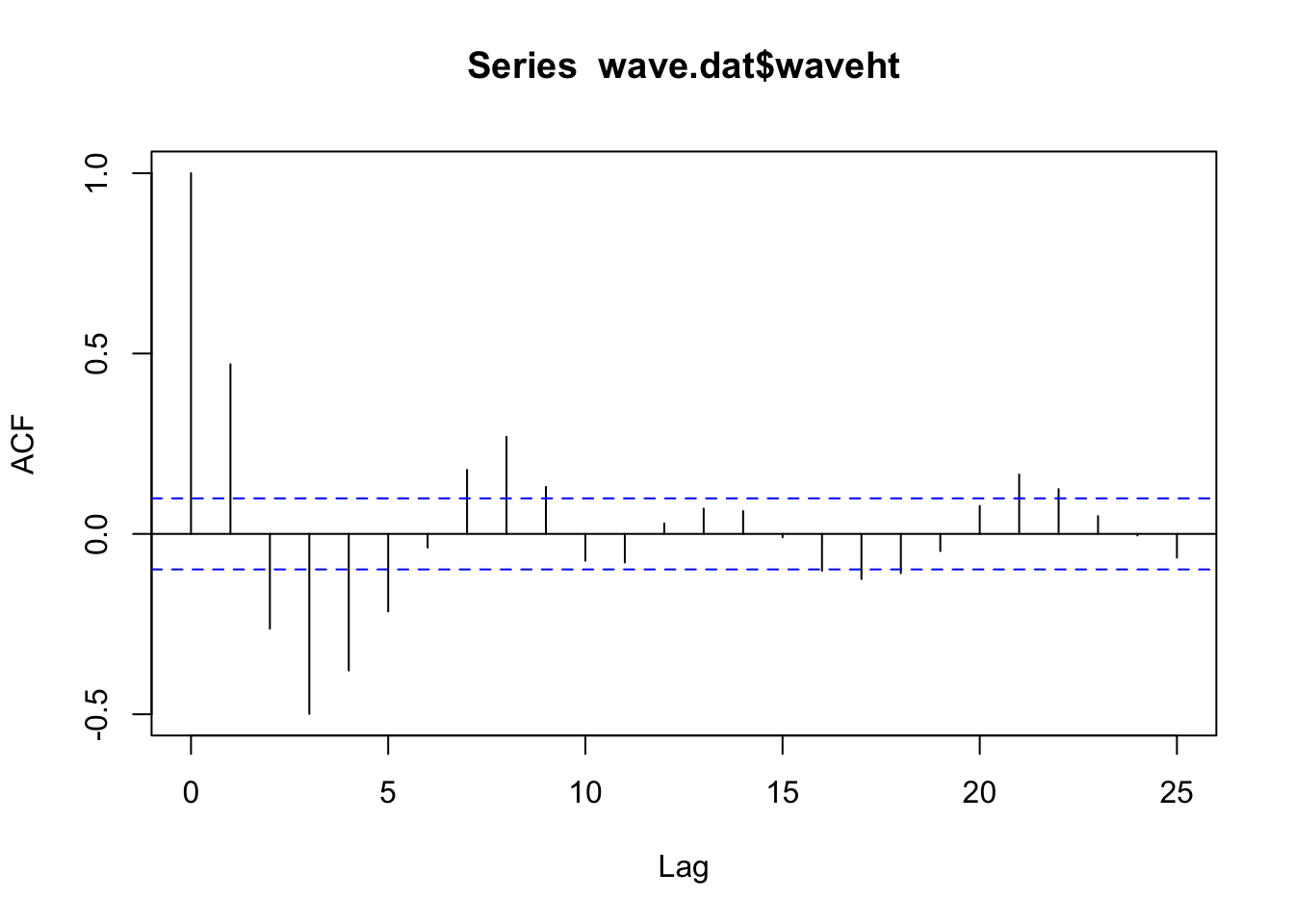
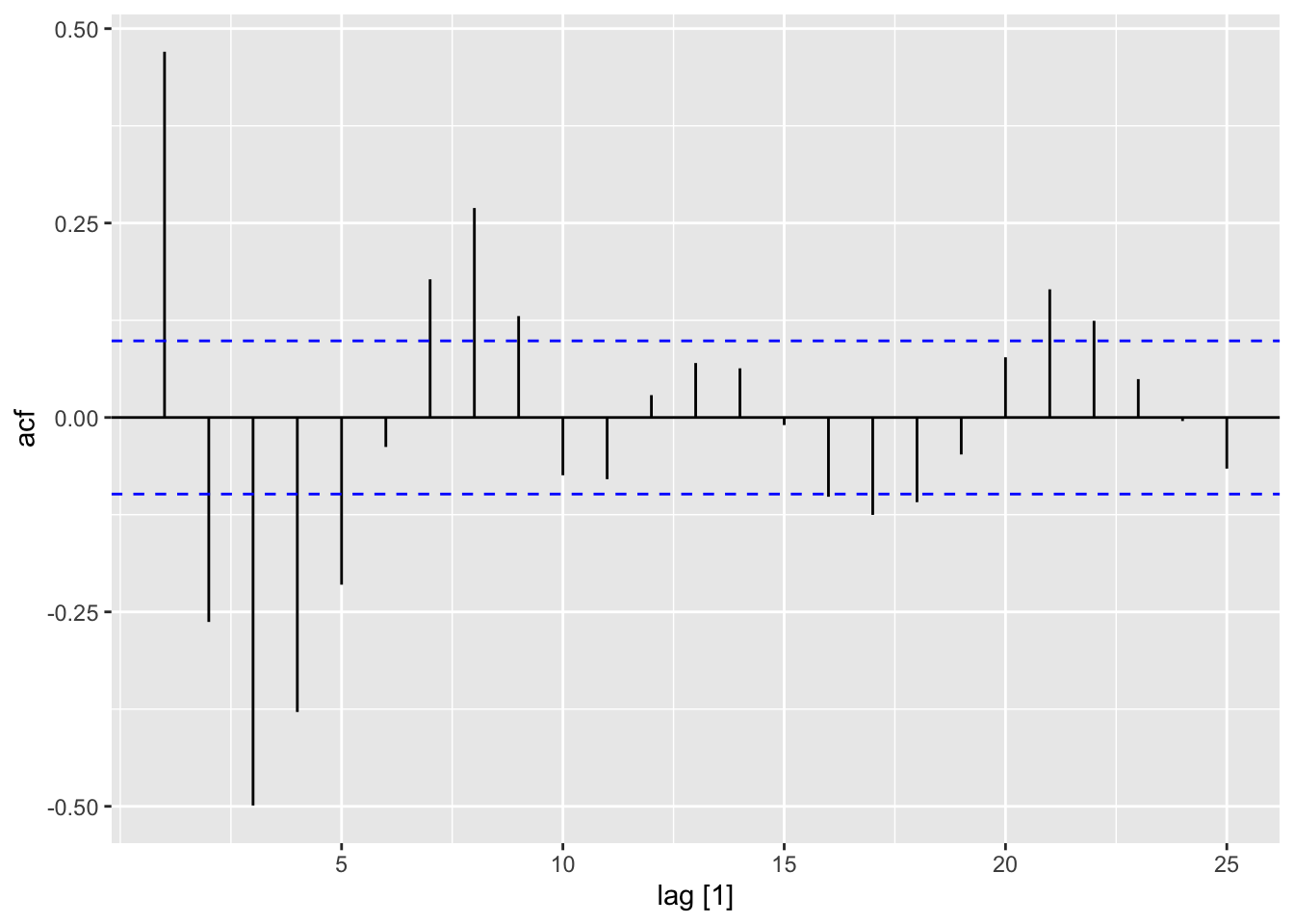
acf (wave.dat$ waveht)$ acf[2 ]
# notice that there is no line at zero ACF (wave_dat) |> autoplot ()
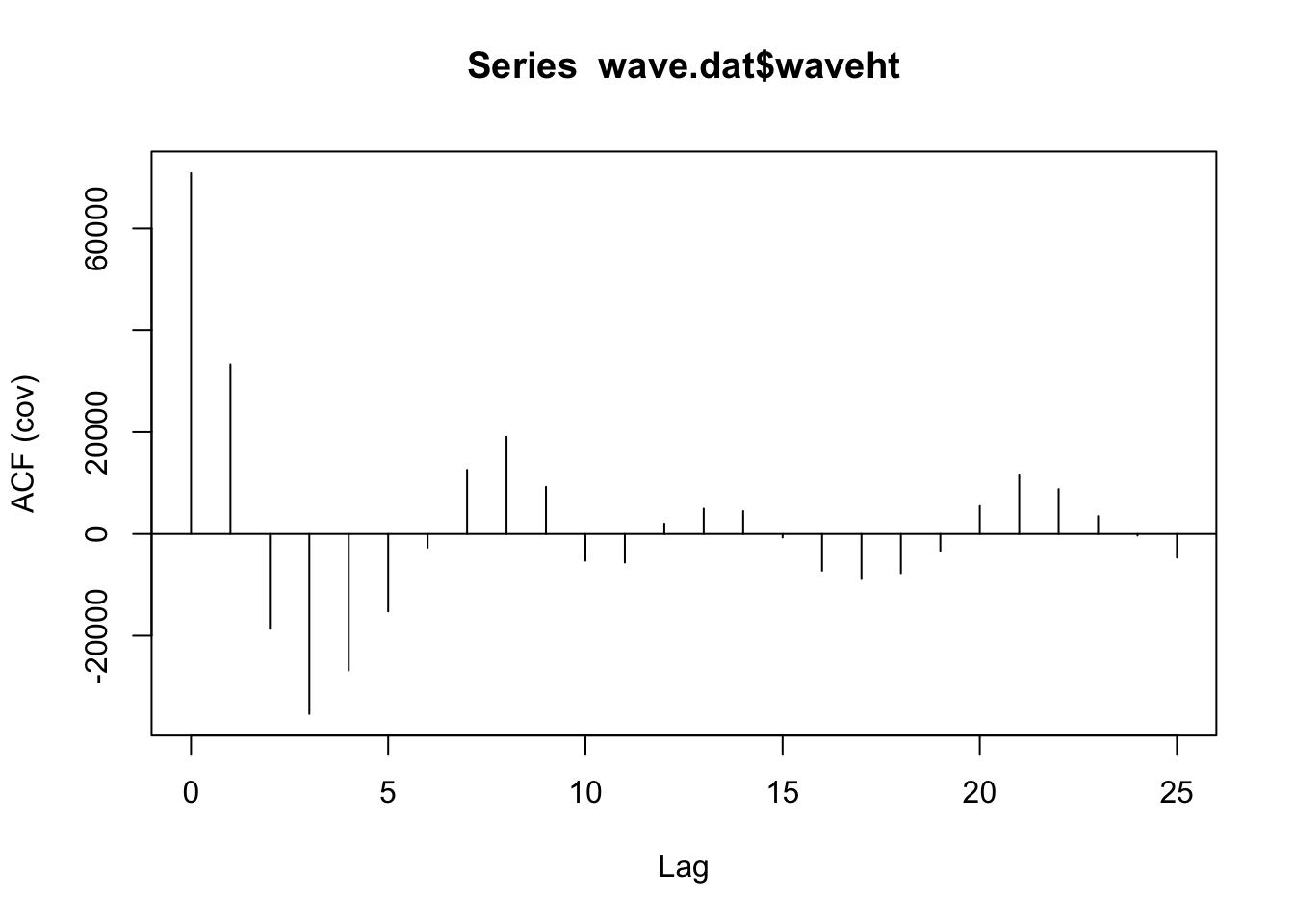
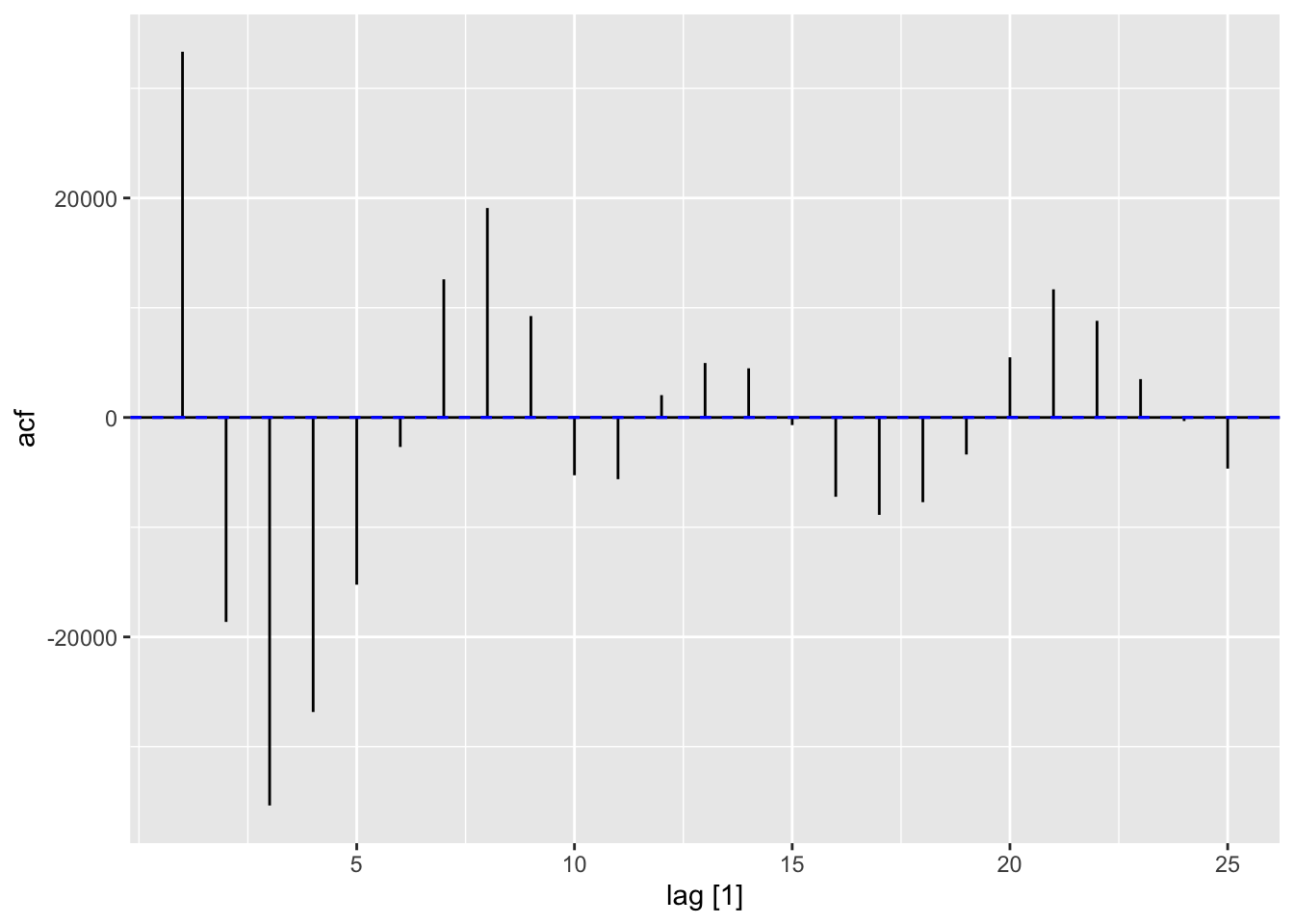
acf (wave.dat$ waveht, type = c ("covariance" ))$ acf[2 ]
ACF (wave_dat, type = "covariance" ) |> autoplot ()
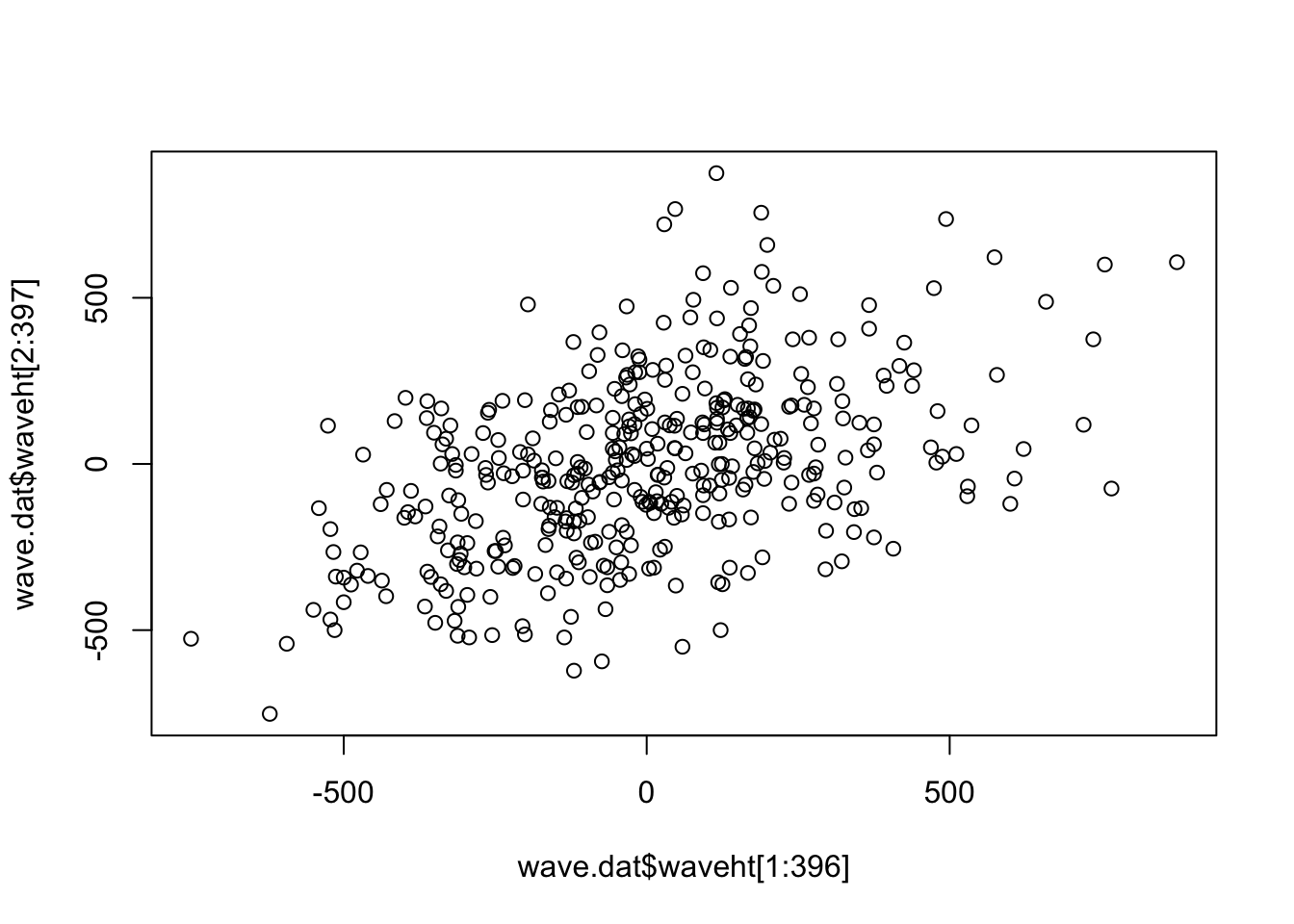
plot (wave.dat$ waveht[1 : 396 ],wave.dat$ waveht[2 : 397 ])
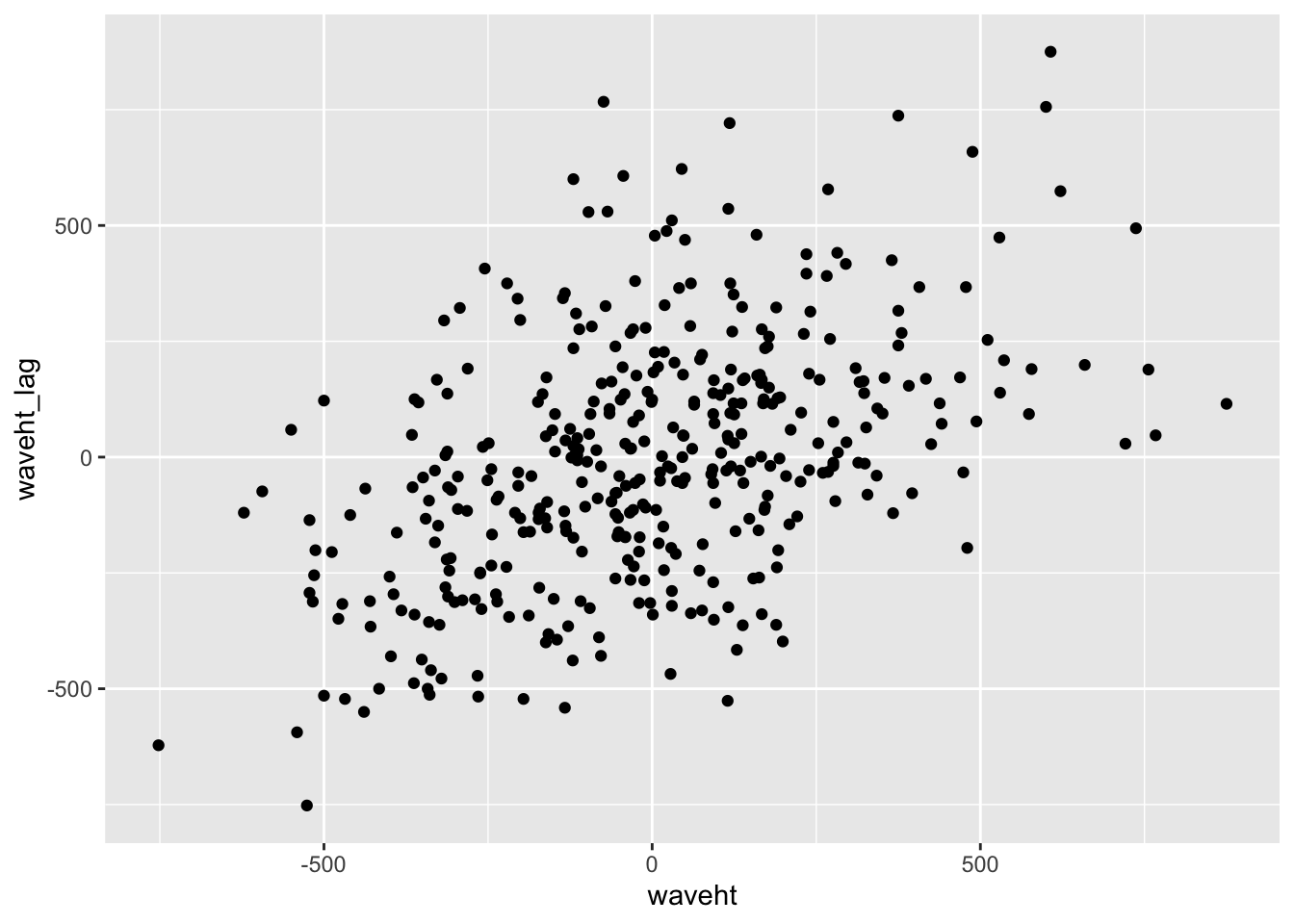
|> mutate (waveht_lag = lag (waveht)) |> ggplot (aes ( x = waveht, y = waveht_lag)) + geom_point ()
Show the Book code in full
<- read.table ("data/wave.dat" , header= T) ; attach (wave.dat)plot (ts (waveht))plot (ts (waveht[1 : 60 ]))acf (waveht)$ acf[2 ]acf (waveht, type = c ("covariance" ))$ acf[2 ]plot (waveht[1 : 396 ],waveht[2 : 397 ])
Show the Tidyverts code in full
<- read_table ("data/wave.dat" ) |> mutate (index = 1 : n ()) |> as_tsibble (index = index)|> autoplot ()|> slice (1 : 60 ) |> autoplot ()ACF (wave_dat) |> autoplot ()ACF (wave_dat, type = "covariance" ) |> autoplot ()|> mutate (waveht_lag = lag (waveht)) |> ggplot (aes ( x = waveht, y = waveht_lag)) + geom_point ()
2.3.1 ‘Correlogram general discussion’ and ‘AirPassengers’ Modern Look
Book code
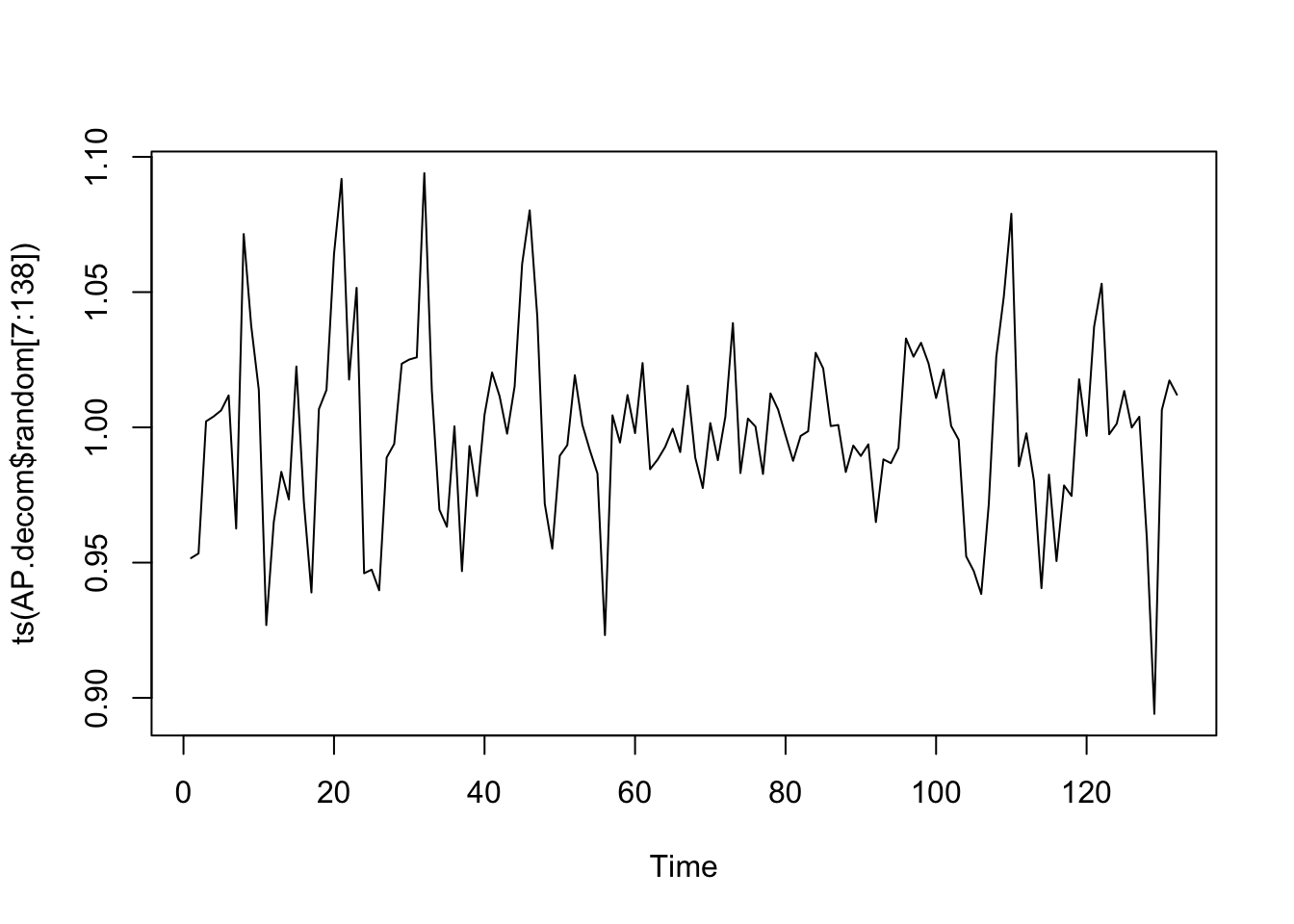
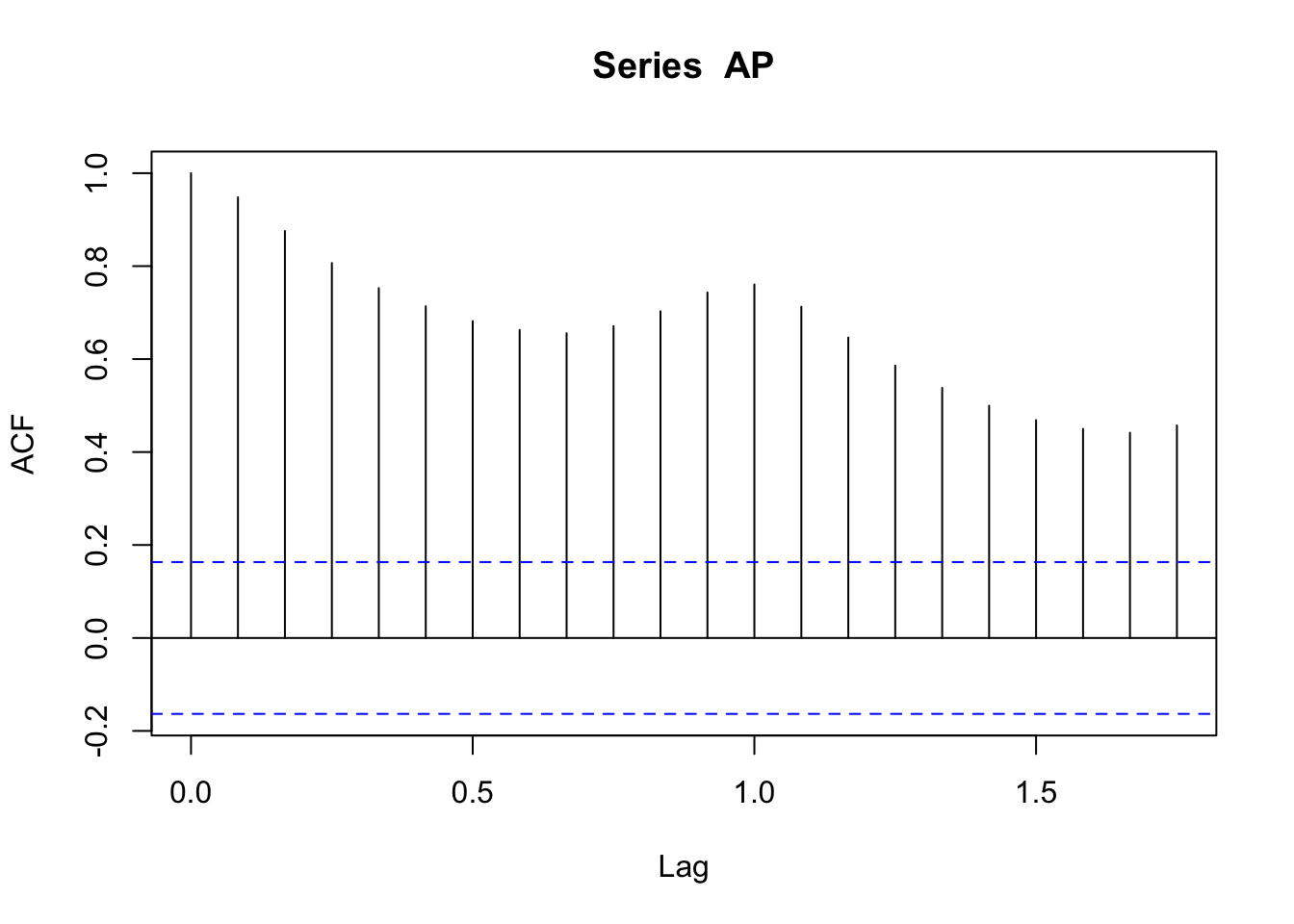
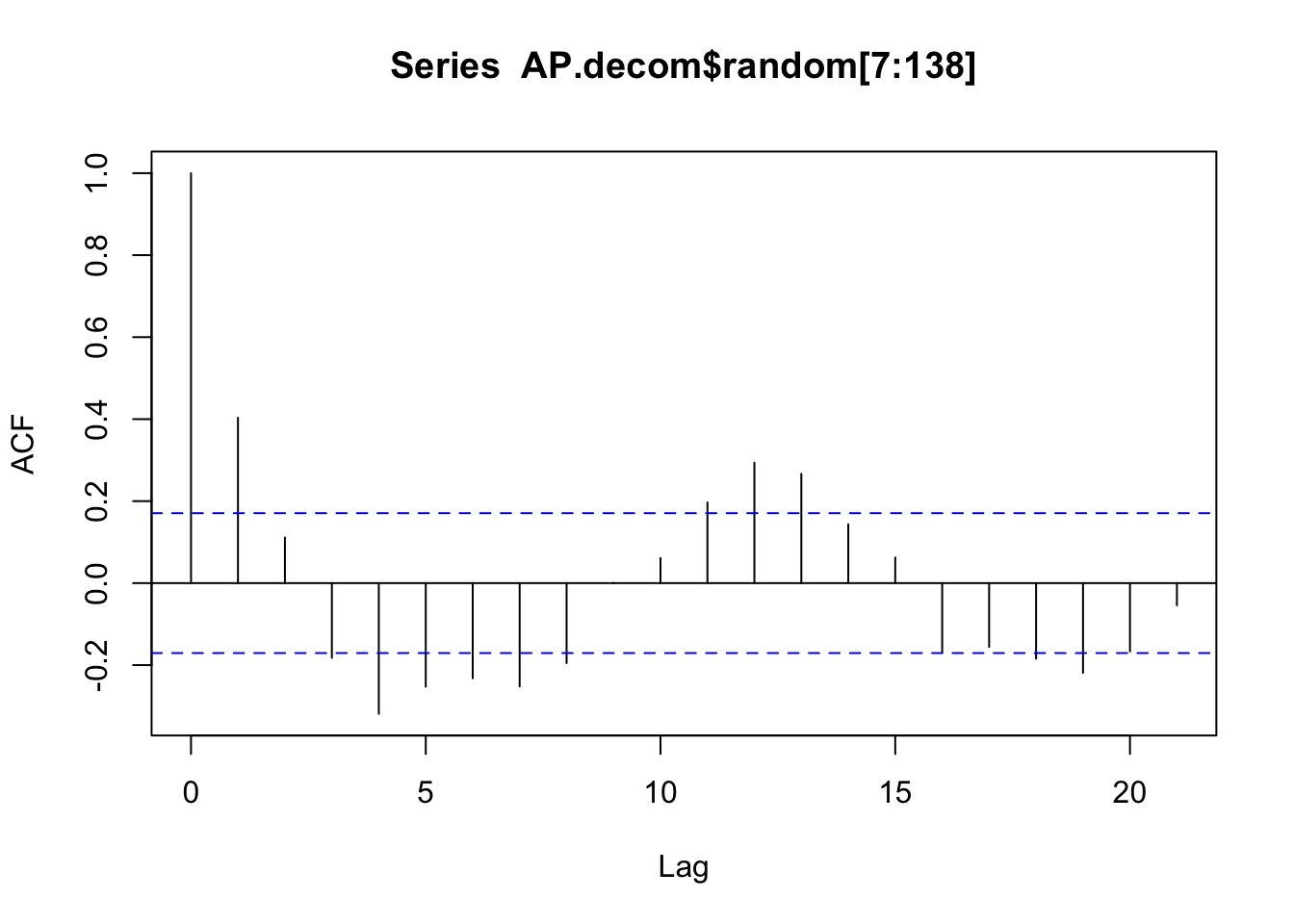
data (AirPassengers)<- AirPassengers<- decompose (AP, "multiplicative" )plot (ts (AP.decom$ random[7 : 138 ]))
Tidyverts code
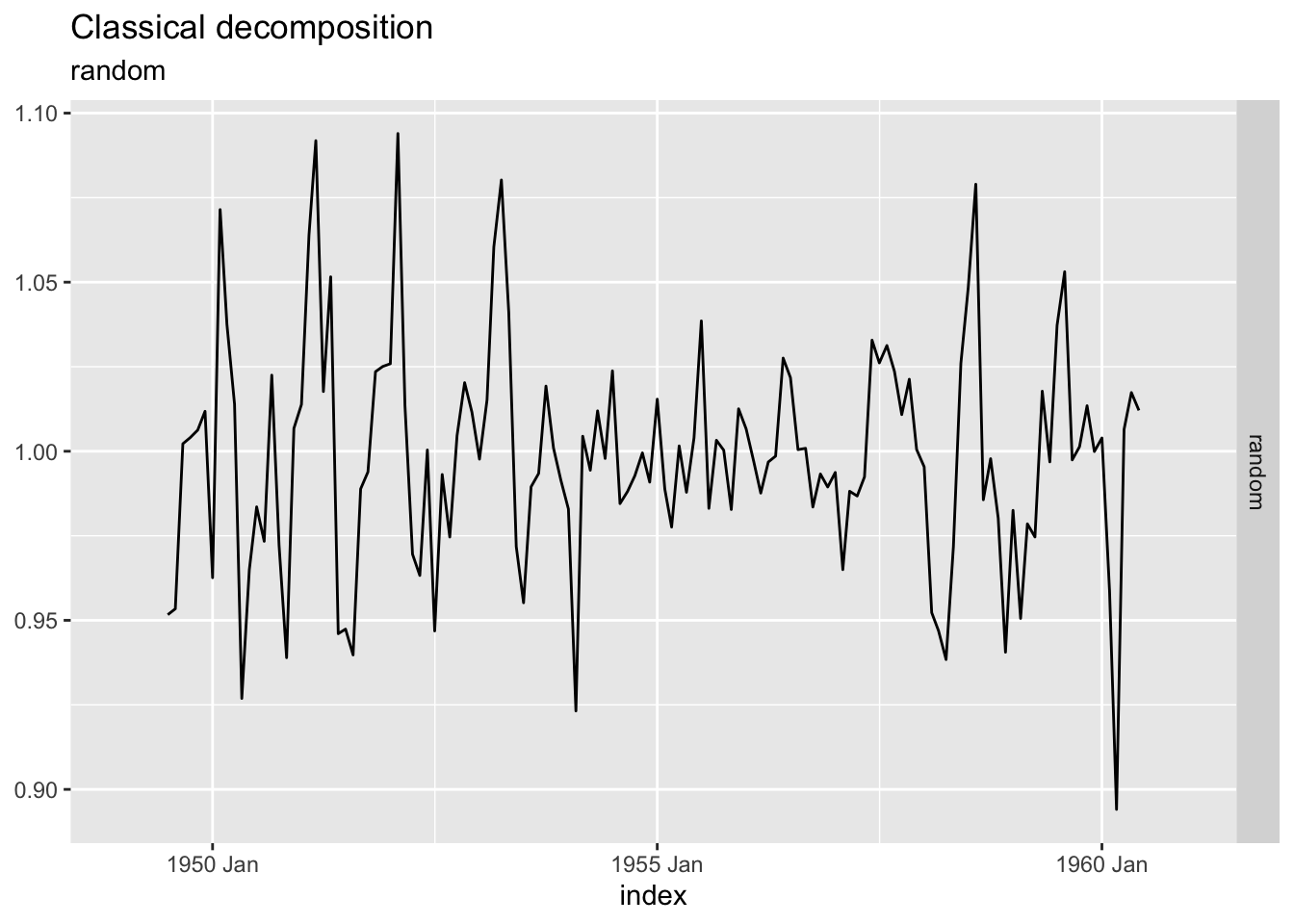
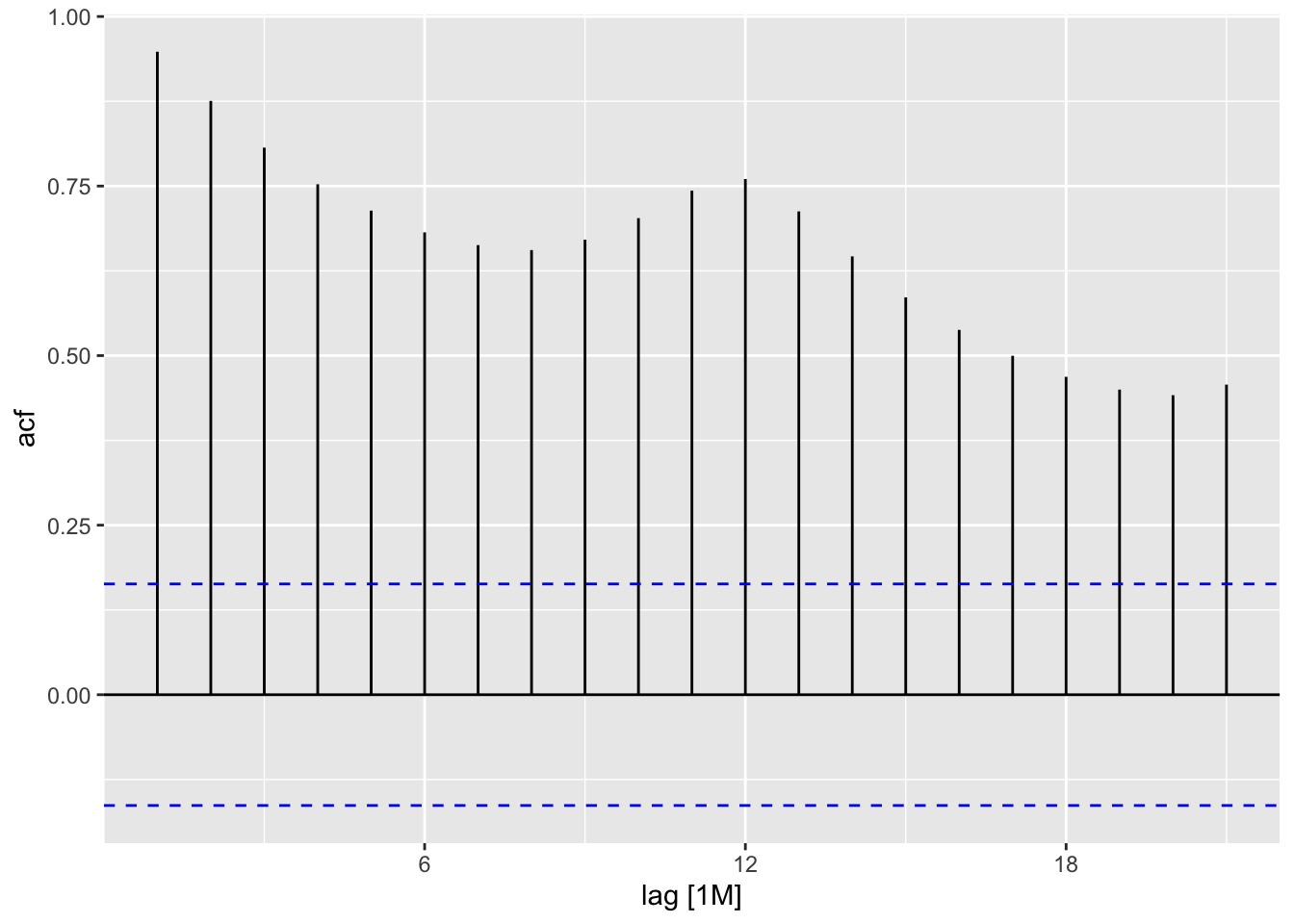
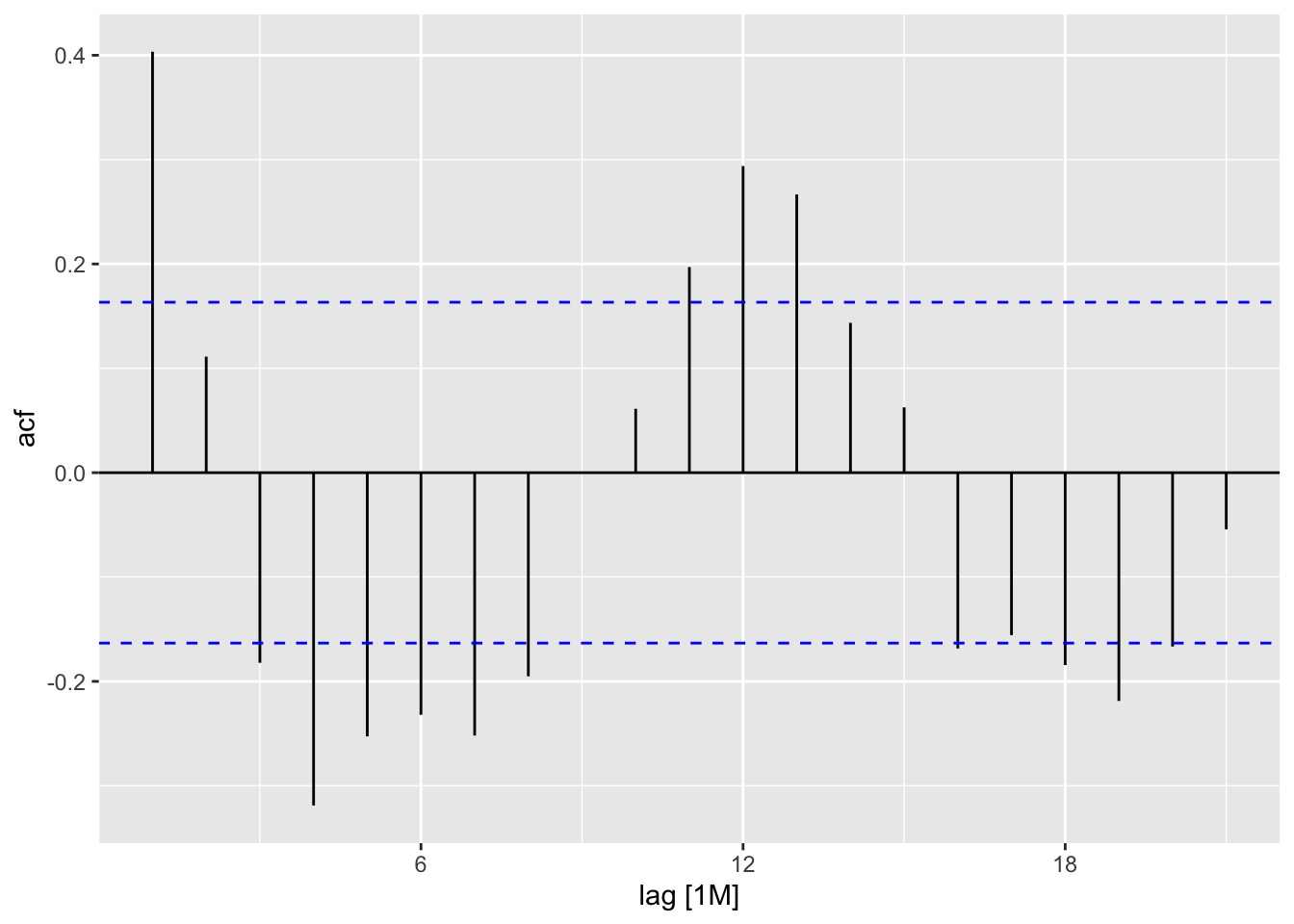
data (AirPassengers)<- as_tsibble (AirPassengers)<- ap |> model (feasts:: classical_decomposition (value, type = "mult" )) |> components ()autoplot (ap_decompose, .vars = random, scale_bars = FALSE )
acf (AP.decom$ random[7 : 138 ])
ACF (ap_decompose, y= random) |> autoplot ()
sd (AP[7 : 138 ] - AP.decom$ trend[7 : 138 ])sd (AP.decom$ random[7 : 138 ])
|> as_tibble () |> slice (8 : 138 ) |> summarise (sd_value = sd (value),sd_mtrend = sd (value - trend),sd_season_adj = sd (random))
# A tibble: 1 × 3
sd_value sd_mtrend sd_season_adj
<dbl> <dbl> <dbl>
1 109. 41.2 0.0333
Show the Book code in full
data (AirPassengers)<- AirPassengers<- decompose (AP, "multiplicative" )plot (ts (AP.decom$ random[7 : 138 ]))acf (AP) # figure 2.6 acf (AP.decom$ random[7 : 138 ])sd (AP[7 : 138 ] - AP.decom$ trend[7 : 138 ])sd (AP.decom$ random[7 : 138 ])
Show the Tidyverts code in full
data (AirPassengers)<- as_tsibble (AirPassengers)<- ap |> model (feasts:: classical_decomposition (value, type = "mult" )) |> components ()autoplot (ap_decompose, .vars = random, scale_bars = FALSE )ACF (ap) |> autoplot ()ACF (ap_decompose, y= random) |> autoplot ()|> as_tibble () |> slice (8 : 138 ) |> summarise (sd_value = sd (value),sd_mtrend = sd (value - trend),sd_season_adj = sd (random))
2.3.3 ‘Font Reservior series’ Modern Look
Book code
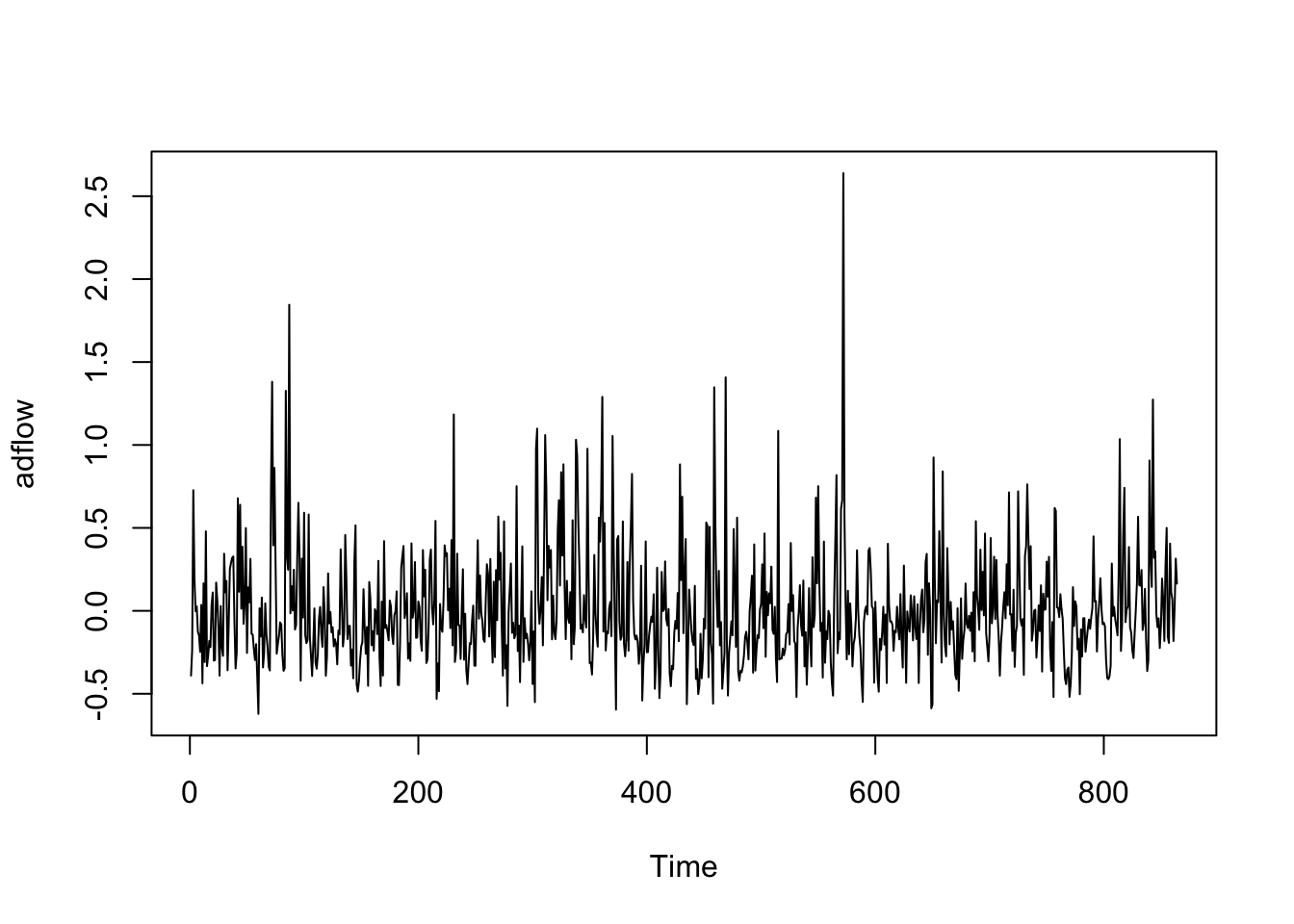
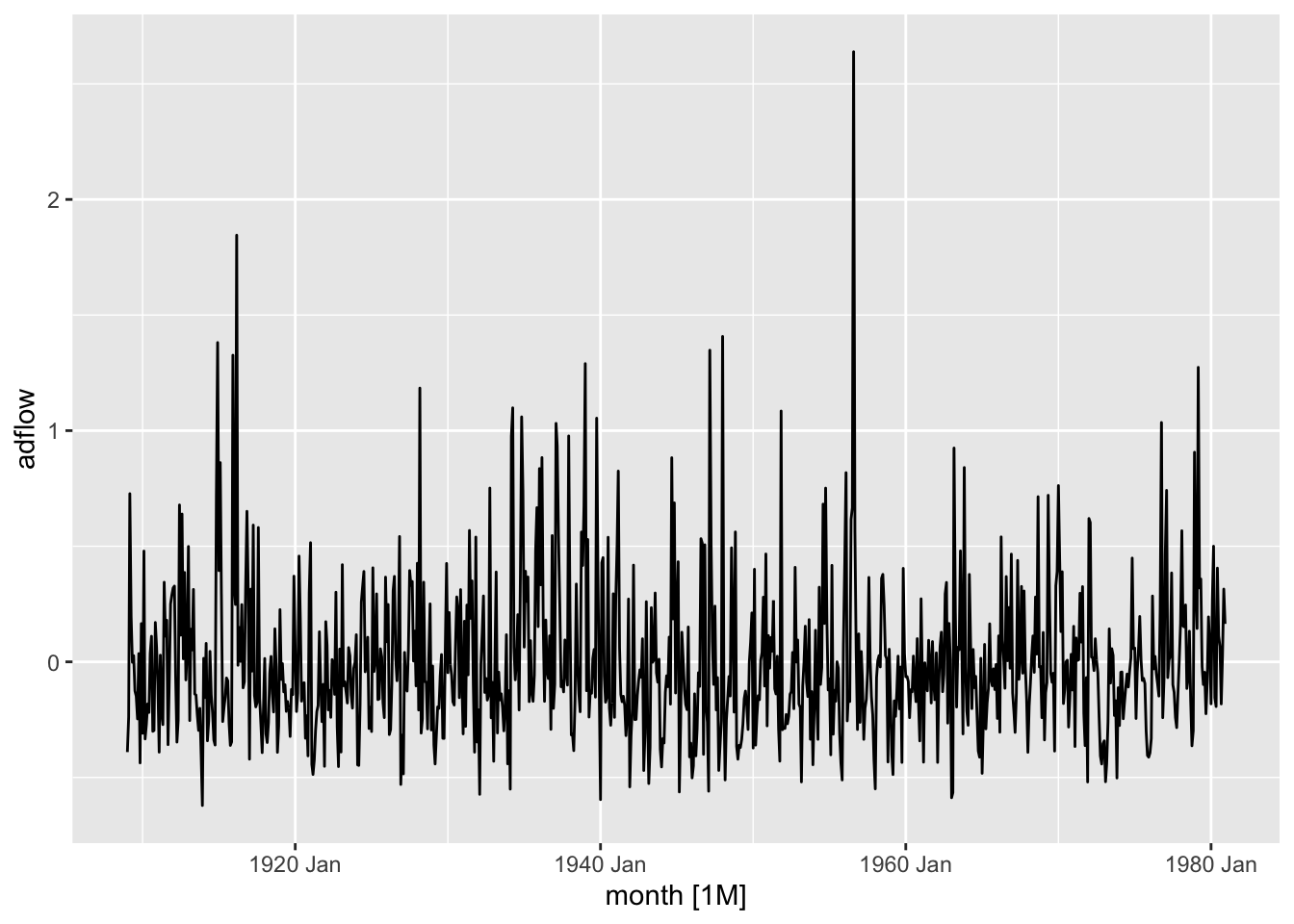
<- read.table ("data/Fontdsdt.dat" , header= T)# attach(Fontdsdt.dat) # bp plot (ts (Fontdsdt.dat$ adflow), ylab = 'adflow' )
Tidyverts code
<- read_table ("data/Fontdsdt.dat" )<- seq (lubridate:: ymd ("1909-01-01" ),by = "1 months" ,length.out = nrow (fontdsdt_dat))<- fontdsdt_dat |> mutate (date = index_dates,month = tsibble:: yearmonth (date)) |> as_tsibble (index = month)autoplot (f_ts) + labs (y = "adflow" )
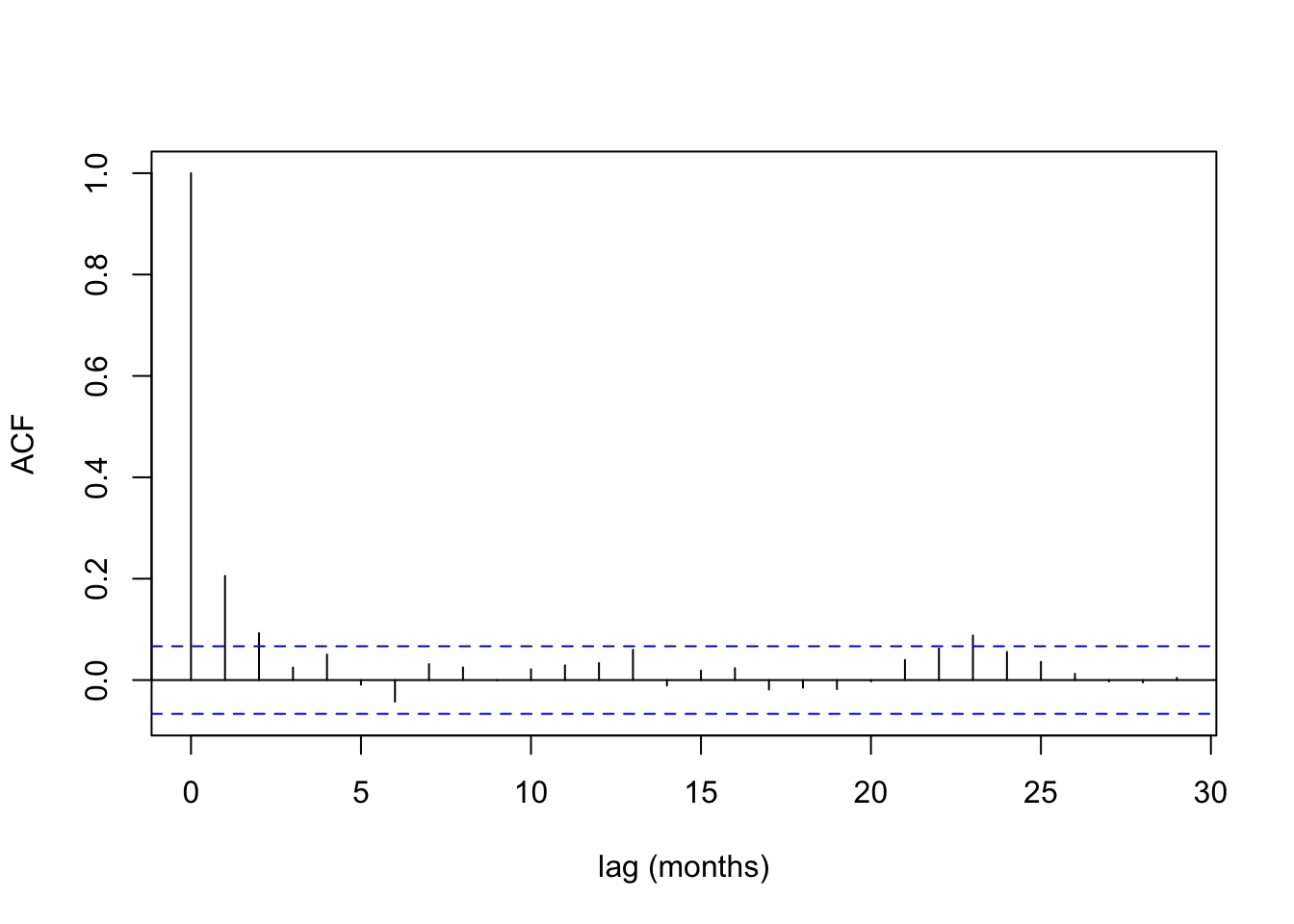
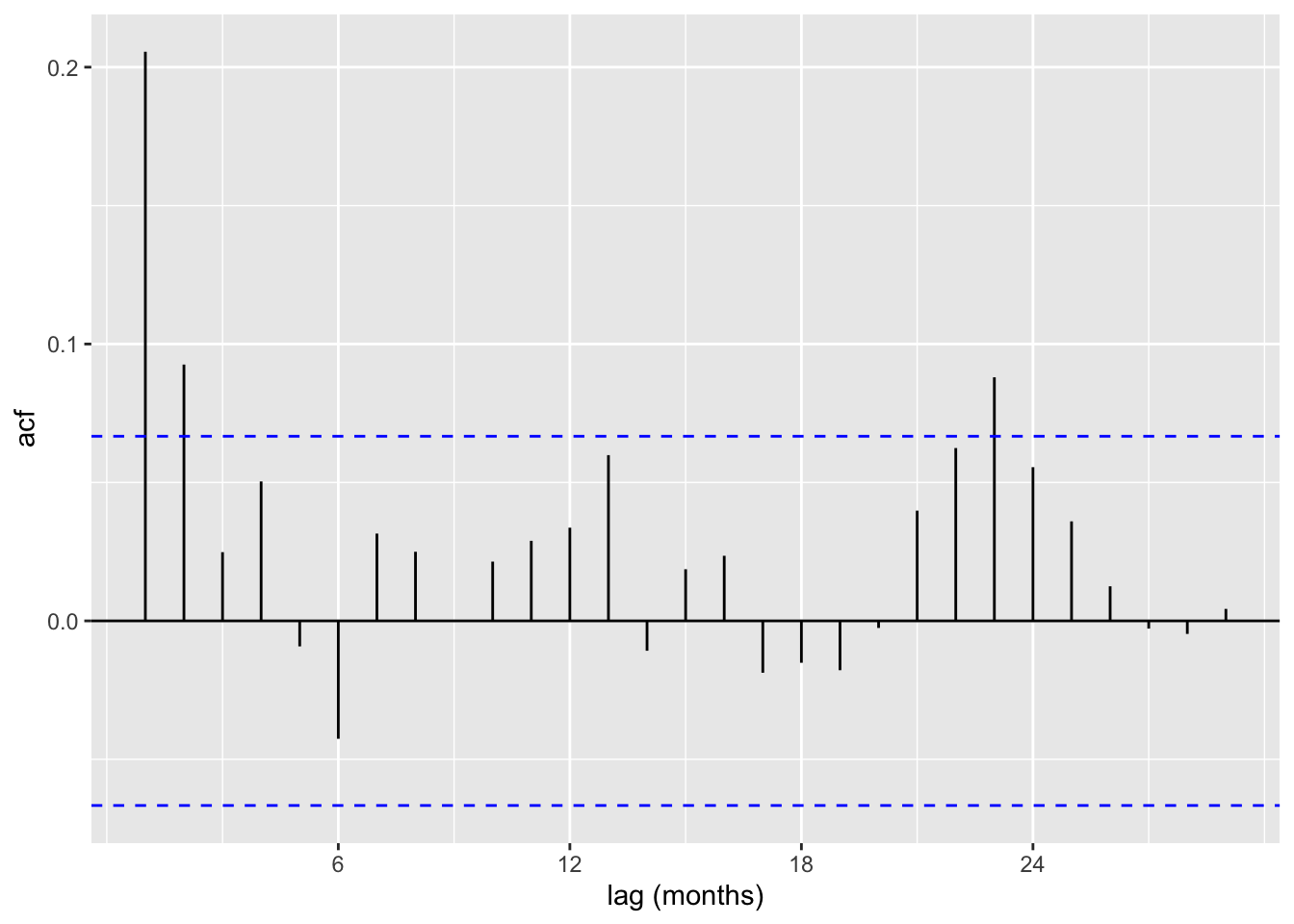
acf (Fontdsdt.dat$ adflow, xlab = 'lag (months)' , main= "" )
autoplot (ACF (f_ts)) + labs (x = "lag (months)" )
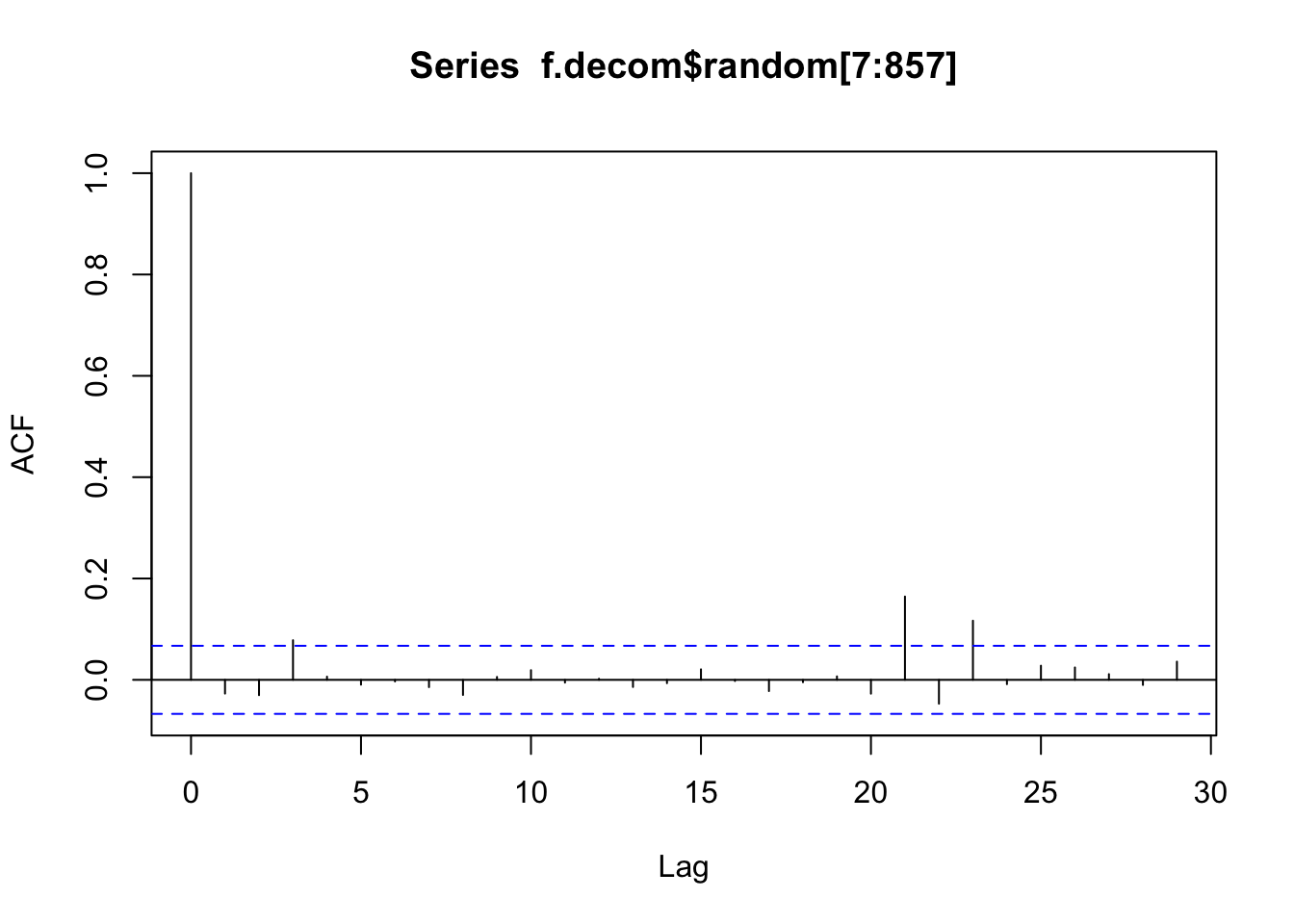
<- ts (Fontdsdt.dat$ adflow, st = 1909 , fr = 12 )<- decompose (f.ts, "multiplicative" )acf (f.decom$ random[7 : 857 ])
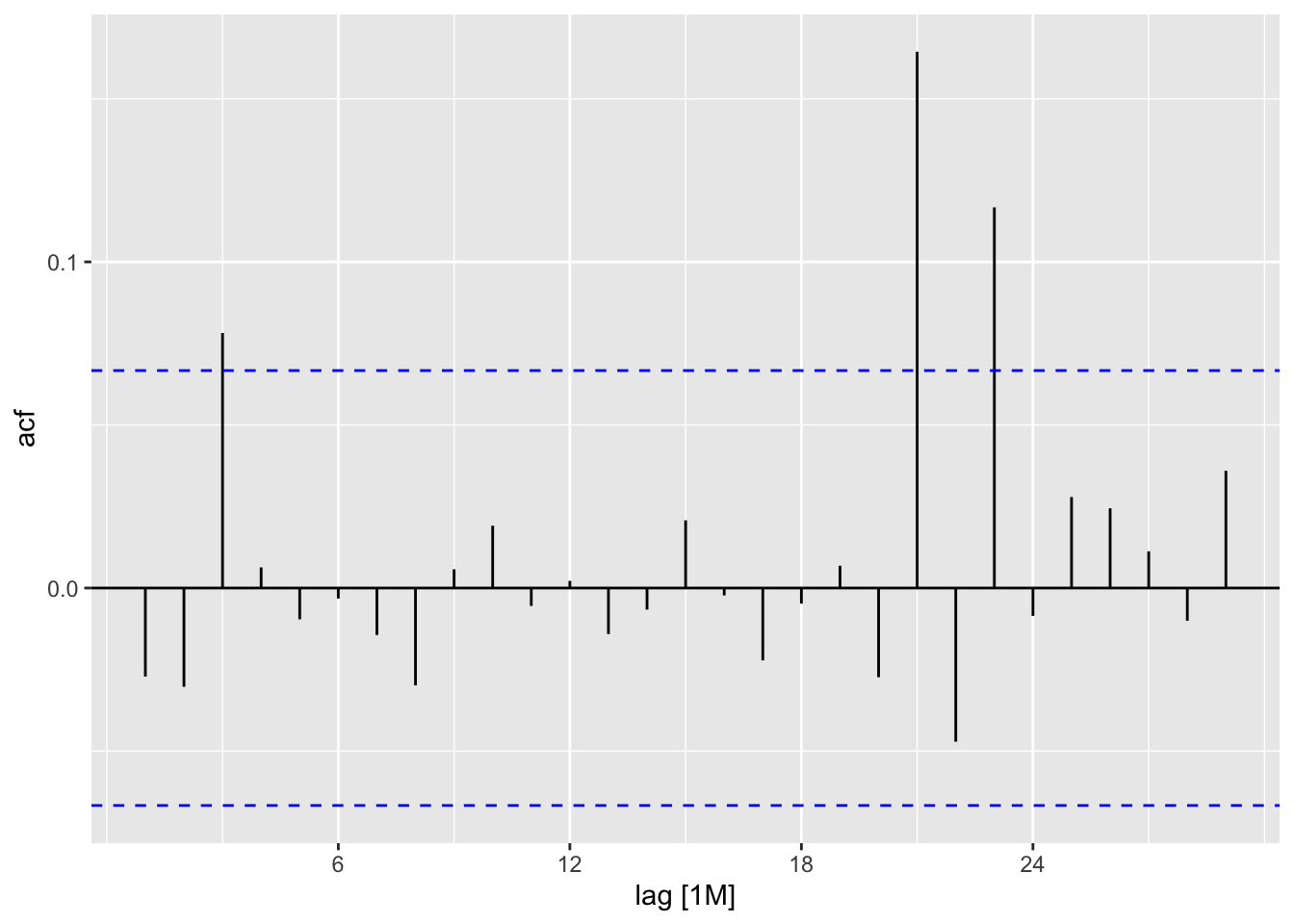
<- f_ts |> model (feasts:: classical_decomposition (adflow, type = "mult" )) |> components ()ACF (f_decom, .vars = random) |> autoplot ()
Show the Book code in full
<- read.table ("data/Fontdsdt.dat" , header= T)# attach(Fontdsdt.dat) # bp plot (ts (Fontdsdt.dat$ adflow), ylab = 'adflow' )acf (Fontdsdt.dat$ adflow, xlab = 'lag (months)' , main= "" )<- ts (Fontdsdt.dat$ adflow, st = 1909 , fr = 12 )<- decompose (f.ts, "multiplicative" )acf (f.decom$ random[7 : 857 ])
Show the Tidyverts code in full
<- read_table ("data/Fontdsdt.dat" )<- seq (lubridate:: ymd ("1909-01-01" ),by = "1 months" ,length.out = nrow (fontdsdt_dat))<- fontdsdt_dat |> mutate (date = index_dates,month = tsibble:: yearmonth (date)) |> as_tsibble (index = month)autoplot (f_ts) + labs (y = "adflow" )autoplot (ACF (f_ts)) + labs (x = "lag (months)" )<- f_ts |> model (feasts:: classical_decomposition (adflow, type = "mult" )) |> components ()ACF (f_decom, .vars = random) |> autoplot ()
2.5 Summary of Modern R commands used in examples