Homework 6.2
1 Ozone
data.dir = "http://media.pearsoncmg.com/cmg/pmmg_mml_shared/mathstatsresources/Akritas"
#8 Age of Trees
oz = read.table(file.path(data.dir,"OzoneData.txt"),header=T)
s_mean = mean(oz$OzoneData)
s_sd = sd(oz$OzoneData)
se = s_sd/sqrt(nrow(oz))
s_mean## [1] 286.3571se## [1] 17.072447 fatty jugs
fat_jugs = c(2.08,2.1,1.81,1.98,1.91,2.06)
#a
sum(fat_jugs>2.05)/length(fat_jugs)## [1] 0.5#b
pnorm(2.05,mean=mean(fat_jugs),sd=sd(fat_jugs),lower.tail = FALSE)## [1] 0.2979415So which is the best value to use?
qqnorm(fat_jugs)
qqline(fat_jugs)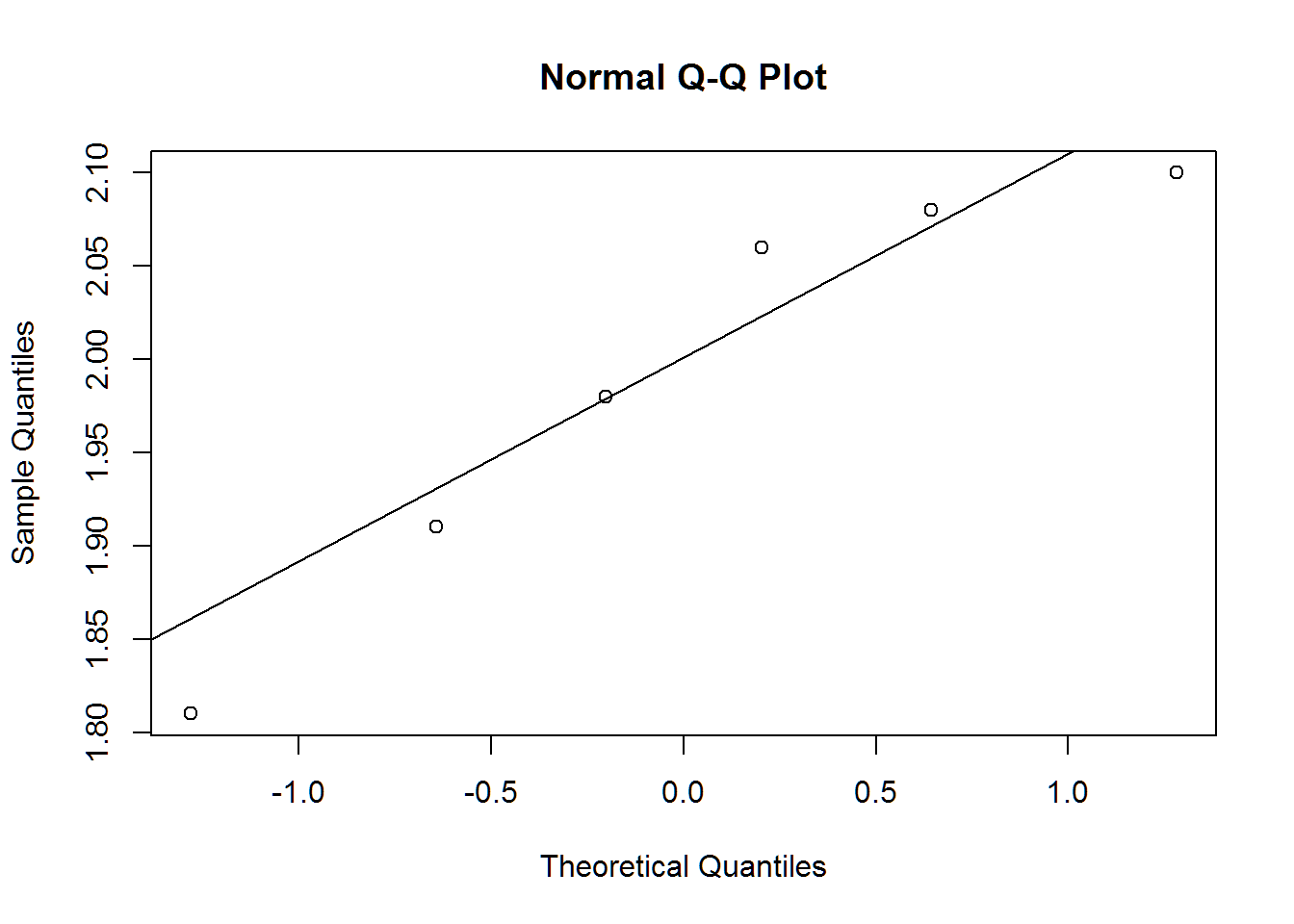
8 Bias in SD
set.seed = 1111
m=matrix(runif(20000),ncol=10000)
var_2 = apply(m,2,var)
sd_2 = apply(m,2,sd)
mean(var_2)## [1] 0.08308021# truth is 0.0833
mean(sd_2)## [1] 0.2345916# truth is 0.2887
# b
m5=matrix(runif(50000),ncol=10000)
var_5 = apply(m5,2,var)
sd_5 = apply(m5,2,sd)
mean(var_5)## [1] 0.08233033# truth is 0.0833
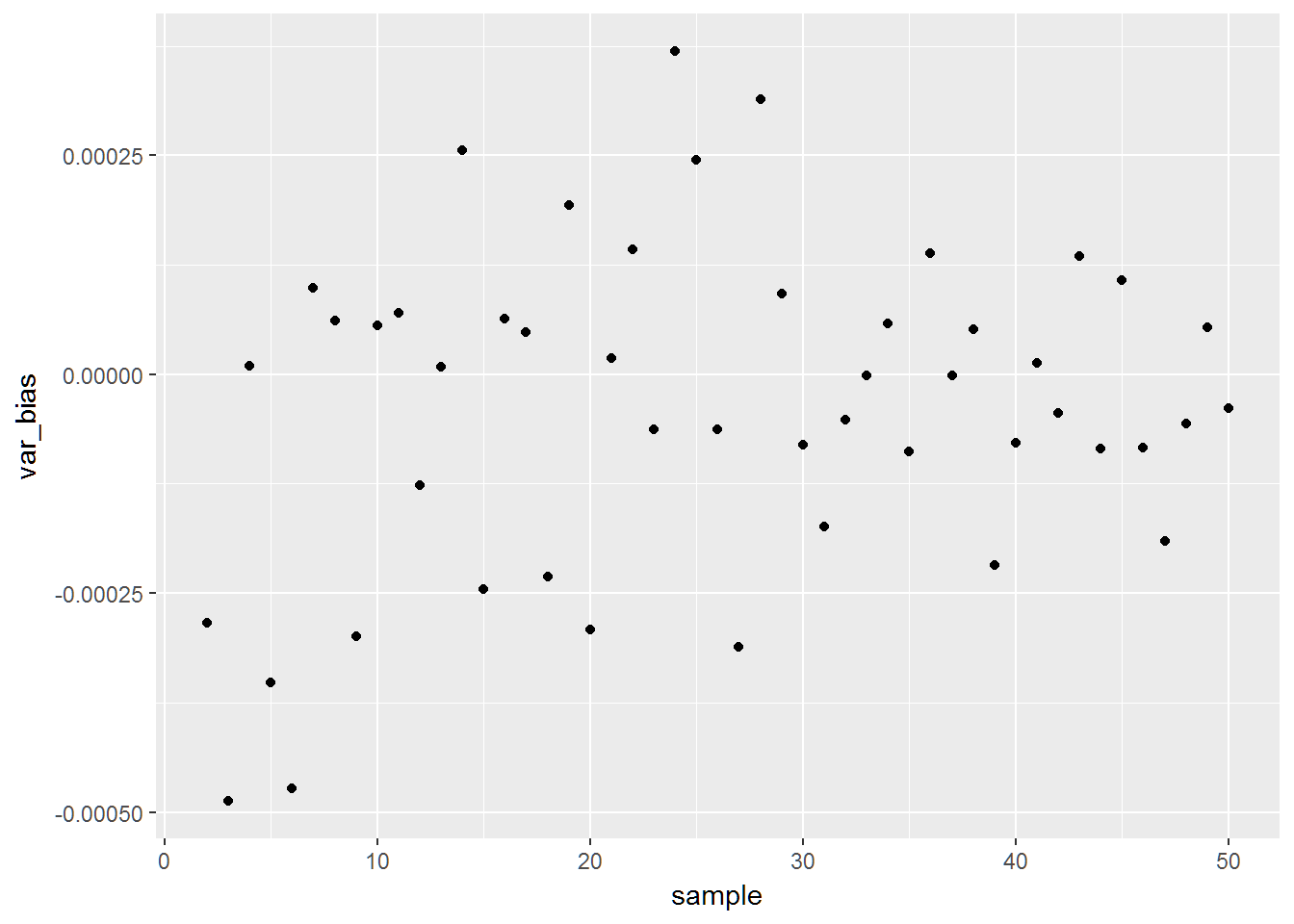
mean(sd_5)## [1] 0.2762757# truth is 0.2887This is interesting. Here is a plot of the bias
#little function to generate data
sd_bias = function(sample_size=2,iters=10000){
out = NULL
for (i in 1:length(sample_size)){
m=matrix(runif(sample_size[i]*iters),ncol=iters)
var1 = apply(m,2,var)
sd1 = apply(m,2,sd)
out = rbind(out,data.frame(sample=sample_size[i],var_bias=mean(var1)-(1/12),sd_bias=mean(sd1)-sqrt(1/12)))
}
out
}
#bias_check = sd_bias(sample_size = 2:50)
#saveRDS(bias_check,file=file.path("data/bias_check.Rds"))
bias_check = readRDS(file=file.path("data/bias_check.Rds"))
library(ggplot2)
qplot(data=bias_check,x=sample,y=sd_bias)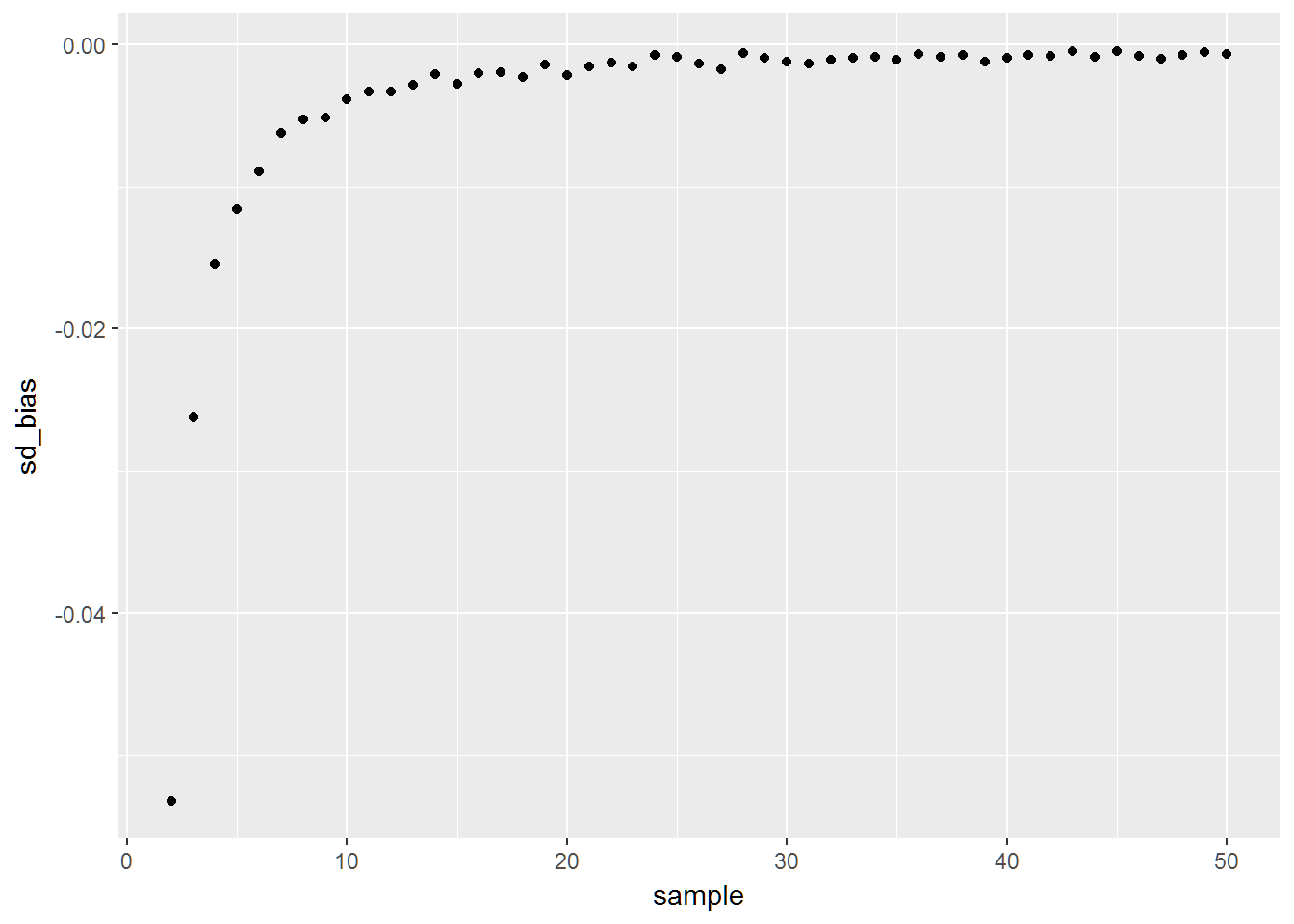
qplot(data=bias_check,x=sample,y=var_bias)